Introduction
With the increase in sequencing activity, and the corresponding increase in virus discovery, plant virology researchers, growers, plant health risk assessors, and plant regulatory authorities have been inundated with the deluge of new virus reports from various virome studies (Massart et al., 2017, Hou et al., 2020, Fontdevila Pareta et al., 2023). The definition of a virome study is open to interpretation but generally includes the application of High Throughput Sequencing (HTS) technologies to describe the virus content from a sample. These sequence datasets may be from individual plant samples as well as from pooled plants in the frame of broader ecological virome studies, or from virus vectors and environmental samples such as soil or water that are directly related to a plant or crop. When looking forward, it is key to consider how the pattern of virus discovery may evolve, because being able to anticipate and adapt to any evolving pattern of virus discovery helps authorities implement timely measures if necessary. Additionally, understanding changing patterns of plant virus discovery can change research priorities and resource allocations. This pattern of discovery is represented in Figure 1.
Exponential phase: As sequencing technology has become cheaper, and approaches have adapted to deal with greater numbers of samples, viromic studies have become greater in scope. Initial virome studies focussed on virus discovery in crop plants and associated non-cultivated species (Coetzee et al., 2010, Fuentes et al., 2013, Gaafar et al., 2020, Adams et al., 2014) to assess the potential risk of hitherto undetected viral infections impeding successful crop production. Since then, there is a trend that sequencing is now increasingly targeting wild plants. Wild plants usually do not display symptoms but may harbour many (still) new-to-science viruses, the sequencing of which has led to a highly productive phase of virus discovery (Robson et al., 2022, Rivarez et al., 2023, Maclot et al., 2020). This increase in reported viruses is accentuated in low resource and geographically remote regions that have few prior baseline plant virome studies, but often greater plant (and therefore plant virus) biodiversity. The exponential phase of plant virus discovery is also accentuated in plant families with a large number of non-commercialised species, such as pasture grasses or weeds as shown recently on temperate pastures and grasslands (Maclot et al., 2023).
Diminishing returns: With an increased global focus on sequencing and virus discovery, eventually diminishing returns are encountered as the likelihood of discovering new viruses decreases, especially on commonly sequenced hosts (Figure 1).
Following the “gold rush” of virus discovery, we might be at the edge of a decline in the number of novel viruses being reported for some crops. For example, on pome and fruit trees, the evolution of virus discovery seems to reach a plateau phase from 2022 after the exponential growth of new virus discoveries between 2011 and 2020 (Figure 2). During the period between 2011 and 2020, the number of viruses identified and known to infect pome and stone fruit trees nearly doubled, increasing from 55 in 2011 to 94 in 2020, with a peak in 2018 when 12 novel viruses were reported. In contrast, since 2020, only five new viruses have been identified, three in 2022, one in 2023, and one in 2024 so far. However, it is possible that the decrease in the rate of novel virus discoveries is linked to the decrease in the sequencing and sampling efforts by the scientific community. More broadly this asymptote may also be asymmetrically influenced by bias in funding streams with greater focus on high value crops (e.g. tomatoes), or on philanthropic funding aimed at low margin subsistence crops (e.g. cassava) and in Low – and Middle- income countries, and focus may then change over time, creating a further bias in data collection. Drivers of virus discovery from changing diagnostic technologies, funding streams, and commodities in trade should also be considered (Fox & Mumford, 2017).
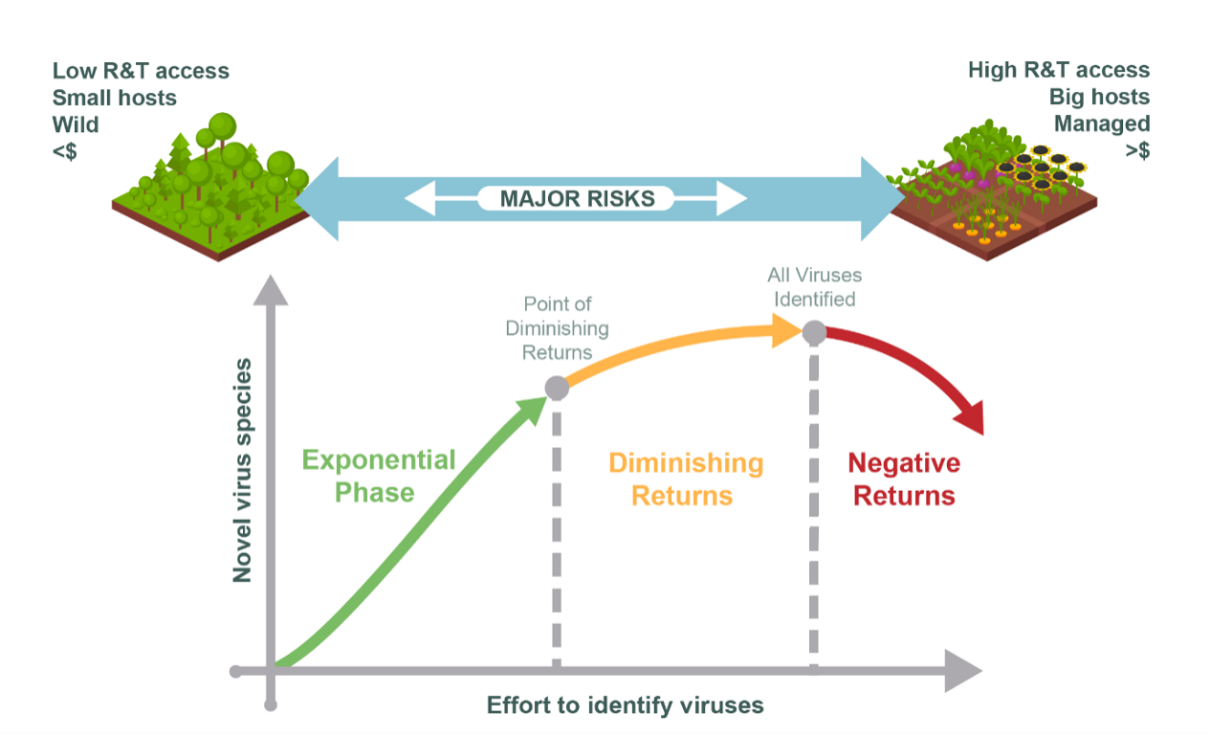
Figure 1 - The pattern of plant virus discovery and risks at the interfaces between extremes of achieved virus identification. Upper: Multiple interfaces and associated unmitigated risks exist between ecosystems with disparate extents of virus identification. The degree of novel virus identification may vary due to research and technology (R&T) access, relative physical size of host plants, degree of ecosystem management, and/or available funds to undertake high throughput sequencing (HTS) of plant viruses. Lower: The pattern of plant virus discovery. The exponential phase generates high numbers of novel plant viruses identified with least effort; diminishing returns of novel virus identification occurs with increased effort until all the plant viruses are described within an ecosystem. Negative returns can occur if effort diminishes the integrity or utilisation of the virus identification process.
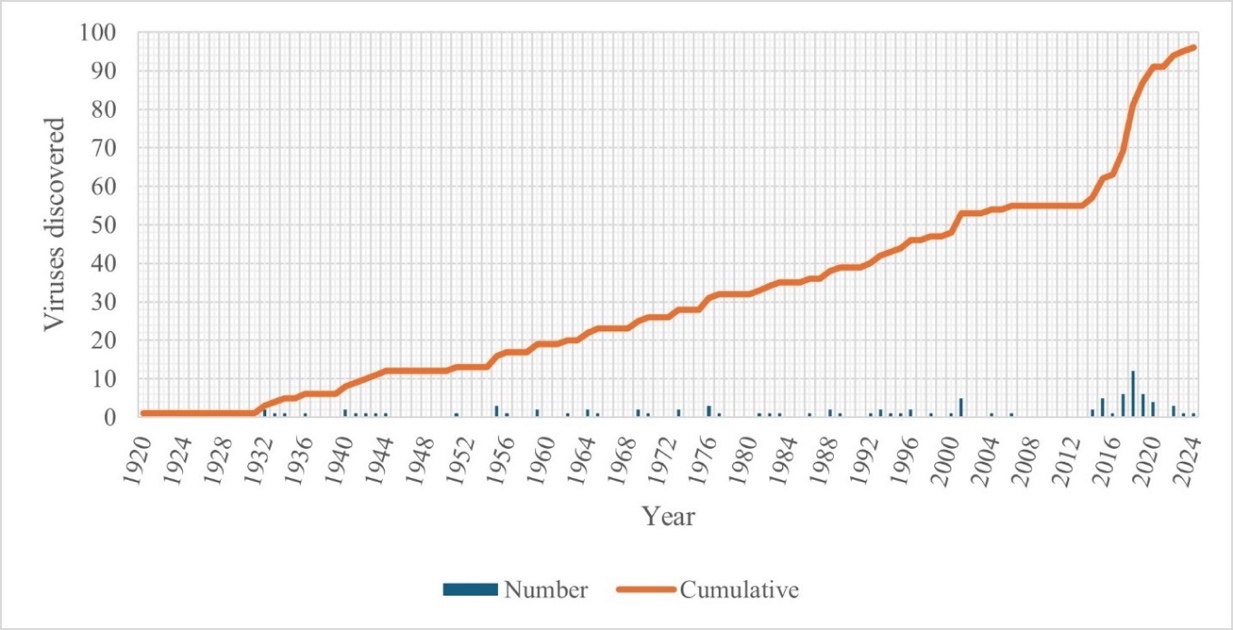
Figure 2 - Evolution of the number and cumulative number of viruses discovered in pome and stone fruit trees. The list of viruses discovered and known to infect pome and stone fruit trees was curated from the available research and scientific publications that were found by a search of online repositories and databases such as PubMed and Scopus. Moreover, these findings were complemented by reference documents, such as the Pest Categorisations from European Food Safety Authority (EFSA), after Fontdevila Pareta (2024) and Fox et al. (2024).
Negative returns: Negative returns are generally produced inadvertently, through lack of resource, knowledge and data sharing. Erroneously annotated genes and genomes within public databases can mislead successive homology-based identification. Multiple identifications and classifications under distinct virus names can also confuse the literature which is clarified through collaboration, re-analysis and publication of synonyms of the virus. Missing sequence data also diminishes the collective returns as data is stored on private hard drives or data ‘clouds’ with restricted access. In this case, access is not restricted due to cultural, social or economic considerations, but usually due to lack of resources or urgency to analyse the data and share the sequences on public databases and journal publications. Efforts to share datasets have been established, e.g. datasharing through the Euphresco network (Euphresco, 2020) Additionally, multiple publications are arising from pre-publication data sharing initiatives (Temple et al., 2024, Alvarez Quinto et al., 2023, Hammond et al., 2021, Kwibuka et al., 2021, Temple et al., 2022), and this has been highlighted as a useful step in handling the deluge of newly discovered pathogens from HTS (Fontdevila Pareta et al., 2023). Regulations become burdensome when they are founded on misinformation. For example, quarantine lists include mandatory testing of phantom diseases and / or disease agents for which there are no sequences/antisera/or no plant reference material available (Tzanetakis et al., 2025). Such phantoms now require formal dispelling through peer-reviewed publication(s) that justify their non-existence and removal from legal processes. However, we are now at risk of the contrary situation of regulations being implemented based on sequence data defining a viral species without supporting biological context.
The biology of viruses within or on plants transiently may generate new identifications that are at first negative returns. Viruses may be moving and replicating in multiple hosts, e.g. both plant and fungal hosts such as cucumber mosaic virus (CMV) and tobacco mosaic virus (TMV) (Mascia & Gallitelli, 2015, Andika et al., 2017), between insects (e.g. rhabdoviruses) (Whitfield et al., 2018), or other organisms. Viruses may be on the phylloplane as virions or in pollen caught in leaf hairs but not infecting the sampled plant (Maclot et al., 2020) or on the seed coat but not infecting the germinated seedling. The ON-plant versus IN-plant assessment may be determined using sample washing prior to virus identification. However, the research questions around biological relevance, i.e. pertaining to infection (present despite washed sample) versus ecology (present without sample washing), can call for distinct and justified protocols. Controls and checks should be included to mitigate possible environmental residue (on or in the plant) or cross-contamination of virus sequences between samples during any step in the process from sample handling to reporting results (Massart et al., 2022). Inclusion of metadata of the process is important when sharing data as this metadata can enable critical interpretation regarding the “ON or IN” virus status of a plant sample. This clarity of process can prevent erroneous reporting in scientific literature, to customers and to authorities. The initial impact of these negative returns, such as erroneous reports, may lead to additional research, new ways of thinking, and technology refinement so it is possible that an initial negative return could lead to a net positive benefit.
Recommendation 1. Pest surveillance in the HTS era: Increased need for baseline surveillance to identify the viral pests present in each country/jurisdiction and determine which pose a plant health risk
MacDiarmid et al. (2013) recommended that As Part of the Burden of Proof, Countries and Jurisdictions Should Identify What Pests Already Exist in, and Which Pests Pose a Risk to, Their Native Flora. In the past decade it has become clear that crops as well as wild plants may be infected by one or several (regulated) viruses, without showing any obvious symptoms (Torrico et al., 2018, Maclot et al., 2020). By focusing on symptomatic crops, latency may go unnoticed until a virus infects a host that induces a visible symptom. The presence of latent regulated viruses does have implications for its (regulated) status. The insight that visual examination of symptoms is not sufficient for the detection of certain viruses may lead to changes in surveillance strategies, as highlighted on pest survey cards from the European Food Safety Authority (EFSA) in the European Union (EU) (EFSA, 2025). In addition to the useful visual symptom-based surveillance, baseline surveillance within the same crop is also advocated, where (bulked) samples are tested irrespective of symptoms (Fowkes et al., 2021, Maclot et al., 2020).
While surveillance and sampling regimes are changing, so are the test methods used for detection and identification of pests in these samples. In the past decade HTS technologies have moved out from being a research support tool and have been used increasingly for plant health diagnostics and virus ecology research (Adams et al., 2018, Villamor et al., 2019, Maree et al., 2018, Massart et al., 2022, Olmos et al., 2018). Guidelines have been written following extensive international collaboration, and these are now publicly available with the aim to facilitate the adoption of HTS technologies and they are now in routine use (Lebas et al., 2022, EPPO/OEPP, 2022, Massart et al., 2022). The combination of different sampling strategies and HTS methodologies has led to an enormous increase in generation of viral sequences and new findings: new for science, new host or new for a territory. Many findings may be unexpected findings, “by-catch”, in plants with or without suspicious virus-induced symptoms, in single plants or in bulked samples, in single or in mixed infection with other viruses.
The increase in reports of plant viral sequences will have impacts on plant health diagnostics, policies and regulation and consequently the trade of plants and plant commodities. Among the findings may be viral sequences associated with regulated pests resulting in phytosanitary measures (Fox et al., 2019, Alvarez Quinto et al., 2023). In order to make science-based, risk analyses, information on biological relevance, epidemiological and contextual information aligned with the viral sequence is required (IPPC, 2019a, IPPC, 2021). However, this supporting metadata is rarely available, or it is often limited in scope or out-dated. Very often, these sequence findings are not confirmed by a second, independent method or infectivity tests, which may also influence the risk assessment of these findings. This knowledge gap is expanding as more new virus findings are reported and the limited resources available to acquire and publish the relevant information cannot match the pace with the speed and diversity of new pathogen findings (Hou et al., 2020).
New plant virus detection/identification technologies
Today there are many new technological opportunities for the detection of plant viruses and determination of their biological impact. These new technologies promise to drive the exponential phase of plant virus discovery and launch us into a new era of high throughput biological insights. The selection of sampling types may be critical for driving plant virus discovery. Using various sampling methods, including samples from various geographic areas and from the wider environment, such as sewage and wastewater, will result in the detection of new occurrences of pathogens (Maksimović et al., 2024, Bačnik et al., 2020). The application of environmental nucleic acid (eNA) sampling approaches for plant virus detection, especially those which utilise HTS based methods, could be used in horticultural drains, which could be considered as the crop equivalents of sewage, or from water courses to monitor the presence of viruses (Nash et al., 2024). There is also the potential in surveying the presence of viruses in dust (of seed lots, from air filters, from spore traps, etc) (Pan et al., 2019), or using insects as sample collectors, either predatory insects or detection from bees and their pollen loads (Fritz et al., 2023, Roberts et al., 2018). Many of these eNA approaches have been applied for targeted testing (e.g. PCR based testing or amplicon based HTS) in the management of viruses such as tomato brown rugose fruit virus, including pollinating insects (Levitzky et al., 2019), from water dispersal (Mehle et al., 2023, Nash et al., 2024), and environmental swabbing (Ehlers et al., 2023, Giesbers et al., 2024). However, when using metagenomic approaches, consideration should be given to the potential sampling bias inherent in these surveillance strategies and to acknowledge that these sampling methods are essentially eNA which is external to host plant. For example, viruses belonging to genera with stable virions are more likely to persist outside host cells in the wider environment, and insect behaviour and feeding preferences will influence which viruses accumulate through trophic levels (Fritz et al., 2023). Consideration should also be given to the nomenclature given to new viruses identified by the eNA surveillance strategies, as described above under “Negative returns”.
Researchers are also exploring existing public (and private) datasets using tools such as Serratus (Edgar et al., 2022), LucaProt (Hou et al., 2024), INFERNAL (Forgia et al., 2023), and vdsearch (Lee et al., 2023). These approaches are being used for expanding the host range and distribution of emerging viruses, viroids, and viroid-like RNAs. This includes expanding the potential host range and distribution of tomato fruit blotch virus (Blouin et al., 2023), for informing the known virome of specific hosts (Rivarez et al., 2021, Rivarez et al., 2023); and for expanding the known viral diversity, such as waikaviruses, secoviruses, and viroid-like RNAs (Sidharthan et al., 2023, Lee et al., 2023, Sidharthan et al., 2024). However, within the plant health community there are increasing concerns around sharing of Sequence Read Archive (SRA) or other raw data on public forums as these can be used for purposes beyond the initial intention (Fontdevila Pareta et al., 2023). Accessing these databases raises issues around privacy within a public arena and specificity of region. Agreements are needed to follow International Plant Protection Convention (IPPC) regulations on what constitutes presence of a species in a region; not just a single record with some or possibly no metadata associated, but evidence to support establishment and active infection of the plant virus in a host plant. The worst-case scenario would be the assumption that the partial genome sequence of a plant virus represents the presence of that plant virus within a sample, or the association of erroneous metadata, such as the location corresponding to the research centre or the sequencing facilities instead of the sampled plant. The verification of the actual host plant should also be carried out by taxonomic assignation of the non-viral sequences in the dataset.
Novel virus identification is also possible using protein homology, e.g. conserved amino acid domains, and using AlphaFold or its next iteration, followed by screening of datasets for recognition of novel virus proteins due to signature similarities e.g. Virtool (Boyes et al., 2020). Such algorithms will likely further develop through artificial intelligence (AI) to increase candidate virus protein discovery, such as in the case of VirHunter (Sukhorukov et al., 2022). Each ‘candidate’ viral protein will then require confirmation using a second, and validated, method of identification, ideally in the context of the host plant, which will also increase the metadata context that might then influence nomenclature. Potentially data sharing and basic supporting research might aid virus identification and will ease the workload of the biological researcher, that is unlikely to be replaced by AI, but will be greatly supported by high throughput mechanisation. However, for the foreseeable future a ”sense check” from expertise will still be needed.
There is an opportunity to predict the impacts of virus infection on individual plants (dependent on concurrent stresses, genotype, species, etc) and the epidemiology of the plant viruses as they spread through or between ecosystems. As AI research and our biological knowledge for plants and their viruses advance, there will be opportunities for high throughput screening/predictions for potential biological impacts of virus infections by analysing uninfected and virus infected (or other types of stress) plants. This AI-based predicted plant virus impact process is likely to slipstream behind technological vanguards based on human virology as it has greatest R&T access worldwide e.g. variants of SARS-CoV-2 (Kosar et al., 2024, Zaib et al., 2021, Yu et al., 2021).
Asymptomatic plant infections
Historically, plant virologists have largely focused on those viruses associated with symptomatic infections, as can be witnessed through virus nomenclature. With the advent of the non-targeted sequencing approaches there has been a deluge of viruses being described without clear association to symptomatic infections. Plants are infected by many viruses without any symptoms as shown by the asymptomatic infection of Arabidopsis latent virus 1 in 25% of the Arabidopsis accessions (Verhoeven et al., 2023) or where the relationship between virus infection and symptomology is unclear, such as viola white distortion associated virus and pansy mottle syndrome (Ciuffo et al., 2014, Fox et al., 2016, Stanković et al., 2021). Additionally, there are examples of virus infections not giving clear foliar symptoms at first, but where there is still a demonstrable impact, such as turnip yellows virus in peas (Nancarrow et al., 2022). Complexities arise when the pathogenicity in a plant host is conditional (Xu et al., 2008, González et al., 2021) and complexity further increases as additional biotic or abiotic interactions are introduced e.g. the tri-trophic interaction of a virus, a fungus and a plant at a hostile temperature (Márquez et al., 2007). In the case of cassava latent virus (CLV), a virus with geminate particle morphology that was detected in both asymptomatic and mosaic-affected cassava plants in East Africa in the 1970s (Bock et al., 1978). Initially, authors dismissed the possibility of CLV being the cause of mosaic disease due to its detection in asymptomatic plants. However, subsequent studies in the 1980s demonstrated that the virus, later named African cassava mosaic virus (ACMV), was indeed associated with geminate particles and cassava mosaic disease (CMD) (Thresh & Cooter, 2005). Further research has revealed the involvement of at least 11 begomovirus species in causing CMD (Legg et al., 2015). Despite this advancement in knowledge, pest lists in many countries include CLV and ACMV, even though the former is irrelevant. This case shows how viruses were listed based on publication records without considering evolutionary knowledge.
Researchers previously blind to many of these virus-host relationships, and are now revealing the plant virome’s complexity including the commonness of asymptomatic hosts. More recently researchers have started to sequence asymptomatic samples from crops and wild plants in the frame of surveys or virome studies (Gaafar et al., 2020, Fowkes et al., 2021, Maclot et al., 2023). These virus ecology studies are also looking at plant virus accumulation in other organisms through trophic levels, e.g. fungi and insects, potentially reporting new virus sequences (Fritz et al., 2023, Forgia et al., 2023). In this way, a new virus species could be identified but without any link to symptoms at the time of discovery or in the case of multi-species pools the host may be unclear, which leaves uncertainty regarding risk. It is likely, in terms of long-term evolution, that all viruses have hosts on which there is no perceptible impact on fitness, e.g., symptoms (Bernardo et al., 2018, Maclot et al., 2020). Therefore, plants may be acting as a “Trojan Horse”, carrying viruses benignly across biosecurity boundaries potentially releasing into new environments, including previously unchallenged symptomatic hosts.
Importantly, plant virus discovery has been technologically constrained due to the small size of virions and the inability to culture these agents outside of a living host. Subsequently, with the advances of sequencing as an unbiased detection tool, diseased samples with an unknown aetiology were interrogated to determine the cause of the symptoms, to validate the causal agent and assess (and perhaps manage) the risk. However, in many cases, complex co-infections, biotic and/or abiotic contributing factors (e.g. temperature or light stress, time from infection, varietal response) were discovered and despite the increased knowledge of virus presence in the symptomatic plant, it still did not allow for a ready understanding of causal associations to symptoms (Fox, 2020). In some cases, the complexity of the causal relationship renders the traditional approach of demonstrating Koch’s postulates inadequate for determining causation in infections with plant viruses. The time required for such biological studies when compared to the relative risks of a new finding may mean that action has to be taken on a precautionary principle e.g. such as the case for ”novel” tymovirus findings in Ullucus tuberosus (Fox et al., 2019, Commission Implementing Regulation (EU), 2018).
Importance of more baseline surveillance crops and wild plants
Although the pace of virus discoveries might be slowing down for some crops, as shown for fruit trees (Figure 2), the asymptote plateau has still not been reached for plant viruses in general. Unless we quantify both the effort and plant virus discovery rates, we do not know where on the curve to pin our current combined knowledge of virus diversity in both crops and wild plants. Only when the virus catalogue is completed can we start to understand the complexity of the virome, the international importance of reservoirs to both crop and wild hosts, and the commonness of asymptomatic hosts. MacDiarmid et al. (2013) highlighted this need for baseline surveillance but still there are very few published catalogues of viral discoveries in different geographic territories (Fox & Mumford, 2017). This lack of rigour is surprising in an age of expanding international trade of seeds, fruit and plants in which each territory has its sovereign right to biosecurity. But true biosecurity requires knowledge of what is present, what is not present, and therefore what is a risk of introduction. It is therefore of increasing imperative that nations gain insight of the virome present within their territory.
Many staple crop plants originate from the Global South (e.g. cassava, taro, potato, tomato, rice maize etc.) (Khoury et al., 2016). While the viruses they harbour may be well described in their new lands of mass propagation, their viromes in their place of origin are less well characterised. Some exceptions are beginning to appear such as potato (Fuentes et al., 2013). A lack of disease epidemics within integrated wild ecosystems, co-evolved mutualism or commensalism, and/or limited resources to identify plant viruses may influence the lack of virus description in the wild relatives of crop plants within their respective centres of biodiversity (Figure 1). Understanding the virome of plants in the ‘Global South’ will likely aid production and provide insights and warnings of new epidemic virus threats for those growing the crops exotically.
Mass monocultures of plants, especially in locations beyond their centre of biodiversity, creates major risks as this provides the perfect opportunity for epidemic events to occur (Figure 1). Such events may occur if a virus enters the monoculture from the new land, or there is a spill-over from indigenous viruses that are local to the exotic place of crop production. For instance, cassava mosaic virus, maize streak virus and cacao swollen shoot virus in Africa are classic examples of new encounter diseases to the crops (cassava, maize and cacao) introduced from Latin America (Kumar et al., 2019). Alternatively, such occurrences may spill-back into the monoculture crop from an incursion originating from the crop’s place of origin but now out of context within a mass monoculture such as wheat streak mosaic virus (Jones et al., 2022), or many of the potato infecting viruses (Santillan et al., 2018, Fuentes et al., 2019, Fuentes et al., 2021a, Fuentes et al., 2021b, Fuentes et al., 2022).
How to do baseline studies
Baseline studies of viruses present within a territory can be undertaken in two distinct ways and should not be confined by the dogma of ‘accepted’ virus host ranges. If the sole aim is to catalogue the viral baseline within a territory then virus reservoir studies may consist of bulked samples (without sample washing). Collection of samples regardless of symptoms is a valid approach in this context to address “what viruses are present?”. Such virome baseline survey is particularly useful to improve the knowledge on geographical spread or host range of known and new viruses and the associated pathways of entry and spread. By contrast, specific surveillance of risk viruses should target known host plants with a focus on high risk/regulated viruses. This need for increased baseline cataloguing of the virome present within a territory is not mutually exclusive from the need to perform targeted surveillance for high-risk plant pests. Plants targeted for collection should be carefully considered with appreciation that the reported virus host range is being regularly expanded, with implications for virus ecology, epidemiology, evolution and impact (Prator et al., 2017, Tollenaere et al., 2016, Moreno & López-Moya, 2020).
Although there is a need for baseline surveillance in different continents both in crops and wild plants it is evident that there is also a need for collecting or receiving relevant contextual data as well as expertise to interpret the relevant information. Early pre-publication data and effort sharing among plant virologists is important, as it can facilitate the process of virus discovery and characterization and reduce the resources and time needed (Fontdevila Pareta et al., 2023). These resourcing factors become even more important in the ‘Global South’ where resourcing has historically been poorer. Inclusion of ‘Global South’ researchers and plant health officers in conferences and workshops enables collegial networks to form and can facilitate the official communications of new viruses in a territory. Such networks can initiate inter-regional projects, and sharing of techniques and data. An example of success is the West African Virus Epidemiology (WAVE) network that has supported the scoping of the cassava/Manihot virome, virus detection and virus removal to transform production from “the grave to great” in sub-Saharan Africa. The Consultative Group on International Agricultural Research (CGIAR) Germplasm Health Units have been surveying for viruses in planting materials of important food staples and related wild species exchanged between the countries, documenting the knowledge on virus occurrence, and developing diagnostic and phytosanitary procedures to generate clean planting materials (Kumar et al., 2021). Similarly, several CGIAR-led initiatives have organised national, regional, and global projects to detect, characterise, and find solutions to economically important viruses (Kreuze et al., 2023, Prasanna et al., 2022).
Recommendation 2: Naming of viruses based on electronic/sequence without context
A decade ago MacDiarmid et al. (2013) recognized the emerging issue of “in silico viruses”, i.e. those viral sequences which were detected through HTS, but not associated with an infection or had limited contextual data. This led to the second recommendation that Plant Virus Sequences Not Associated with a Recognized Virus Infection Are Designated as “Uncultured Virus” and Tentatively Named Using the Host Plant Species of Greatest Known Prevalence, a General Location Identifier, and a Serial Number. Here it is important to differentiate between the virus as a physical entity and the species concept as a taxonomic construct (Van Regenmortel, 2007). In the intervening decade there have been developments with the International Committee on the Taxonomy of Viruses (ICTV) to address some of the confusions with virus nomenclature, and move to a binomial species classification and nomenclature approach for ratified genera and species (Siddell et al., 2020). Although this move was not without disagreement from within the community (Gibbs, 2020), the move to binomial species nomenclature was ratified for virology and was completed for currently ratified species by 2023 (Walker et al., 2021). The ICTV publishes demarcation criteria for Genera and Species which include multiple characteristics including biology (vector, hosts) and genome organisation. Despite this, the determination of tentative novel species, especially from metagenomic data, is initially performed by the determination of percentage-identity to characterised species. This approach has allowed the rapid molecular classification and initial taxonomic placement of thousands of newly detected viruses in the sequencing era, to the point that not only novel species, but also novel genera have been ratified only by the genomic sequence, e.g. Genus Chordovirus (Adams et al., 2014, Adams & Kreuze, 2015, Svanella-Dumas et al., 2018, Fox et al., 2022a). However, the ICTV are very clear that they do not determine “virus names” only those of species.
The ICTV has now completed the three-year task for the study groups to assign a binomial-format name for ratified species. One aim of this move was to more clearly delineate between “official” species names and “vernacular” virus names. Recommendations here are therefore intended to deal with the vernacular naming viruses, not species nomenclature, which is the domain of the ICTV. The aim is to help with avoiding confusion regarding the large numbers of viruses which are being described from metagenomic sequence data, often with little biological context as to their origins or impacts. Whilst the ICTV recognise that metagenomic viruses, i.e. those without supporting biological context, could be incorporated into official classification schemes (Simmonds et al., 2017), this has potential implications for the regulatory authorities, beyond the practical consequences of renaming hundreds of virus names as synonyms to ensure compatibility across multiple databases (e.g. UK plant health Risk Register, EPPO Global Database, CABI Crop Compendium).
The challenges posed by findings of “electronic” metagenomic viruses without biological context, and the deluge of new virus discoveries facilitated by the application of HTS, were discussed by MacDiarmid et al. (2013). However, the additional complication of thousands of SRA-inferred viruses being described, detected outside the confines of their original project scope with minimal contextual data, had not been anticipated. Frameworks for dealing with this increase in novel findings have been produced for determining the potential biological impact and risk associated with the discoveries (Fontdevila Pareta et al., 2023), but have thus far avoided suggesting harmonising conventions on virus nomenclature. Plant viruses (as well as species) need clear and unambiguous naming conventions, not least because biosecurity authorities need to regularly assess the impact of emerging virus threats.
At the core of plant health risk assessment is the need to have a “characterised” pest (IPPC, 2019a, IPPC, 2021, IPPC, 2019b), and this process is greatly aided by unambiguous naming. Trying to link disease with “causal agent” and then multiple potential pathogen names can lead to confusion and potentially erroneous regulation. Therefore, the essential recommendation is that vernacular virus nomenclature needs to be clear and unambiguous, with each viral entity having a unique and clearly identifiable “label” synonymous with the official species assignment. As one of the drivers for the ICTV moving to a binomial system was to help clarify the difference between the official and the vernacular, this should be at the core of virus naming
The ICTV policy to move to a binomial system for species will potentially address Recommendation 2 proposed by MacDiarmid et al. (2013) to tentatively name those viral sequences that have been detected by HTS. Reference to the provisional taxonomic placement (genus) should be included where possible when using the vernacular virus nomenclature. Additional information in the virus name, such as reference to biological context (host and disease associations) might be included as discussed below. Where possible adhering to the traditional “time served” convention for virus names would be desirable, namely a format of “Host – symptom – virus” e.g. cucumber mosaic virus, but with reference to the genus, i.e. cucumber mosaic cucumovirus. The additional use of the taxonomic placement in the virus nomenclature has been previously applied, and would allow for an easier read across from vernacular to species e.g. tomato spotted wilt orthotospovirus. Additionally, continuing the historically recognised use of letter-based nomenclature, initiated after Kenneth Manley Smith described potato virus Y and potato virus X as distinct entities (Smith, 1931b, Smith, 1931a), should also be honoured for future findings in genera recognised to have this naming convention. Though extending this practice outside of these historical conventions should be avoided.
There are however, two potential issues with a diseased/symptom based convention for novel viruses identified based on genome sequence alone. Firstly, the lack of knowledge of biological context for many new findings (Hou et al., 2020) will make this challenging without further work which could take several years. This also raises discussions on the difficulties of definitively linking pathogen to disease by use of biological experimentation (“Koch’s postulates”) (Fox, 2020). One solution for viruses identified via metagenomic approaches, or those with an uncertain link to disease causality, would be to adopt a format “Host – genus – index number” e.g. carrot torradovirus 1 (Adams et al., 2014). However, as previously mentioned in recommendation 1, many viruses are discovered through either virome and environmental monitoring studies or increasingly data mining, and host related naming conventions for such viruses will not be appropriate. The term “associated” is increasingly being used for both viruses where there is not direct causal link to symptoms and those viruses detected inadvertently in non-plant studies. Due to the potential for confusion to arise, “associated” should only be used where there is suspicion of a novel virus being found in an infected host. Where a plant virus is detected through SRA searching or through trophic accumulation studies from an organism which is not thought to be a host, then the word “derived” is proposed to be used instead. The other issue which arises in novel virus discovery will be those viruses being described which taxonomically fall outside of currently recognised viral genera. Here we recommend either assigning at the family level, or if phylogenetic evidence supports creation of a novel genus, then this should be proposed e.g. Chordovirus (Adams et al., 2014).
To aid with delineating between “virus name” and “species name”, we also recommend researchers to suggest both a vernacular name and a tentative species name in a publications e.g. parsley umbravirus 1 (PaUV-1) and Umbravirus petroselinum (Fox et al., 2022b). Whilst the ratification of species names remains the preserve of the ICTV, it is suggested that a way of handling non-ratified species names, could be to add a non-italicized “sp.nov.”, or “sp.” as suggested for uncultivated virus genome sequences (Adriaenssens et al., 2023a, Adriaenssens et al., 2023b), after the proposed species name to show it has not yet been formally adopted, as in bacterial and plant taxonomy. Researchers should also be encouraged to submit an accompanying virus taxonomy proposal to the ICTV on reporting the discovery of previously undescribed virus taxa.
Recommendation 3. Biosecurity risk assessment of plant viruses requires investment in time, new technologies and ways of working to determine the biological impact of plant viruses
Previously, the third recommendation recognised the potential growing gap between ease of discovery and the challenge in generating biological knowledge of these newly described viruses. This led to the third recommendation to Invest in Basic Research to Determine the Ecology of Known and New Viruses with Existing and Potential New Plant Hosts and Vectors and Develop Host-Virus Pathogenicity Prediction Tools (MacDiarmid et al., 2013). The potential for erroneous or overly rigorous application of regulations, or even deliberate application with malicious intent to create a trade barrier, is an area which creates concern in the application of HTS, and the publication of pathogen presence, in many countries/regions. Strict adherence to the International Standards on Phytosanitary Measures (ISPMs) and fulfilling the first and third recommendations from MacDiarmid et al. (2013), to ensure that a country is surveying for the presence of a pest, and that it is characterised before implementing a pest risk assessment (PRA) and regulation, should help allay some of these concerns. The risk assessment of a plant pathogen is carried out to standards administered by national and regional plant protection organisations (NPPOs and RPPOs) and coordinated by the IPPC. These ISPMs give set standard approaches for assessing the risk of emerging plant pests. Before a pest is assessed to have regulatory status, such as being declared a quarantine pest, there is a formal Pest Risk Analysis (PRA) process (IPPC, 2019a, IPPC, 2021, IPPC, 2019b). Among the key criteria considered in this risk assessment is the characterisation of the pest, including its taxonomic placement, distribution, host range, transmission mechanisms, and the likely impact on crops and the wider environment. There may be a reaction to a novel discovery in one territory which provokes implementing trade restrictions in another, without the necessary data to support a PRA being generated.
To assist with supporting the risk assessment of new viruses discovered through HTS, guidelines have been published to suggest a scientific and regulatory framework in which to operate (Fontdevila Pareta et al., 2023). Whilst some pest categorisation has been conducted by EFSA within the EU region, viruses associated with limited sequence or biological characterisation should not be included within EU regulation e.g. non-EU viruses of Fragaria, Cydonia, Malus and Pyrus (Bragard et al., 2019b, Bragard et al., 2019a).
The number and rate of viruses being described, and ratified as species, has increased over time (Figure 3). The move from “biological“ to molecular diagnostic characterisation can be divided into the pre-sequencing era where virus diagnostics was primarily carried out with bioassay, electron microscopy and serology, and the post-sequencing era where initially Sanger sequencing and latterly HTS methods are predominant (Fox & Mumford, 2017, Jones et al., 2021). The boundary between these era’s is hazy but the key milestones are the first sequence of a plant virus generated by Covey et al. (1981) and the first demonstration of HTS in a plant virology context (Adams et al., 2009, Kreuze et al., 2009, Al Rwahnih et al., 2009). This change in the application of methodological approaches means that there is a growing gulf between the new viruses being reported and the knowledge of their biology and consequent risk. This is highlighted by Hou et al. (2020) as an emerging biological desert. Addressing these challenges are of particular importance to improve biosecurity risk assessments, reducing the risk uncertainties, especially for new virus species.
One application where the massively multiplex detection potential of HTS has been slow to be fully exploited is in the area of post-(and pre-)entry quarantine screening (Lebas et al., 2022). Whilst the relatively slow processing times for samples makes it unsuitable for testing at the border, in some globally traded commodity crops, high grade breeding material may be subjected to exhaustive virus testing under licence prior to release. This is routine practice in clonally reproduced crops such as potato, citrus, stone fruit trees (e.g. plum, peach, almond), and soft fruit seedlings, such as strawberries. As this screening covers a range of viruses for which, historically, diagnostic tests were unreliable in propagation plants, the testing involves biological screening/indexing as well as molecular and serological testing. In most cases the testing can take multiple years to complete a full cycle of testing (Maree et al., 2018). Rather than replacing these traditional approaches, HTS has initially been used to supplement the testing, pre-selecting those breeding lines most likely to fail further screening and allowing for early intervention. The Australian and New Zealand Plant Post Entry quarantine programs have adopted HTS to improve quarantine testing by detecting all regulated exotic viruses from imported plants in a single assay (Gauthier et al., 2022, Delmiglio et al., 2023, Whattam et al., 2021).
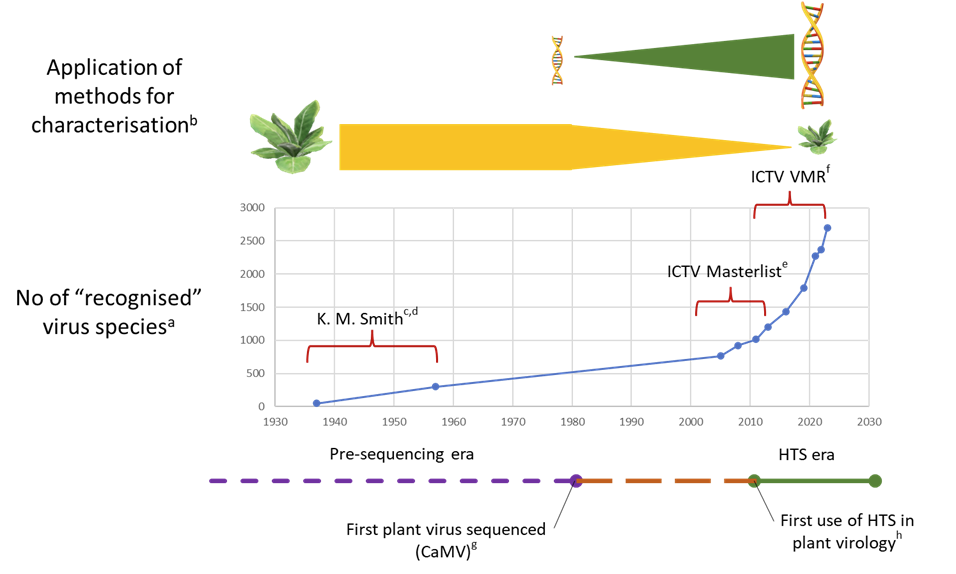
Figure 3 - The relationship between the application of diagnostic technologies and the discovery of newly described viruses. (a) The method for capturing the number of virus species has changed over time. The earliest records included here are from K.M. Smith and predate the formation of the International Committee on the Naming of Viruses (ICNV) the forerunner of the ICTV in 1966 and the virus species concept (Smith, 1937, Smith, 1957); (b) Application of biological vs molecular characterisation based on Fox and Mumford (2017) (c) after Smith (1937) (d) after Smith (1957) (e) ICTV Master species lists 2005-2016 (ICTV, 2024a) (f) ICTV Virus metadata resource (VMR) 2017-2023 (ICTV, 2024b) (g) First sequence of a plant virus (CaMV) published (Covey et al., 1981) (h) First publications demonstrating the use of high throughput sequencing in plant virology (Adams et al., 2009, Kreuze et al., 2009, Al Rwahnih et al., 2009)
There are practical challenges to the implementation of a single diagnostic test for all possible viruses in a sample, such as what plant material to sample, and when, to give the optimal chance of detection of a virus (Malapi-Wight et al., 2021); what depth of sequencing is needed to allow for confidence in detection sensitivity (Visser et al., 2016, Pecman et al., 2017); and side by side validation of the new method with the gold standard in line with international standard methodology (EPPO/OEPP, 2018b). However, a key question raised by applying HTS in this way is how to redefine “freedom from…” in plant health regulation, where some commodity crops may harbour persistent lifestyle viruses and benign infections. For example nearly all sweet potatoes are infected with badnaviruses, that seem not to affect the plants in any detectable way (Kreuze et al., 2020) and likewise many pepper varieties and some bean varieties are infected with endornaviruses without any apparent symptoms or negative effects (Okada et al., 2011, Okada et al., 2013, Okada et al., 2017). CGIAR genebanks and breeding programs have adopted pre-export screening of clonal germplasm with HTS as a risk prevention measure (Kumar et al., 2021). However, the discovery of cryptic virus genomes in some materials complicates the export decisions due to the lack of clear guidelines regarding declarations and processing information for import decisions.
The other challenge in this area are the historical listings of “phantom viral agents” without supporting molecular characterisation data, which are included in listings. With the move over the last 20 years to the broad application of target-specific molecular diagnostic tools in plant health (Fox & Mumford, 2017), there is no way to readily identify these agents. Without historical reference isolates, there is also no guarantee that these agents have not been rediscovered and characterised under a different name, as happened in the case of plantain virus X and Actinidia virus X (Hammond et al., 2021). More than 130 such phantom agents, across 10 host genera, have been identified in a recent review (Tzanetakis et al., 2025). These are listed as part of certification and plant propagation schemes across the world, but with a diminishing access to the skills and funding needed to run post-entry quarantine programmes with biological indexing, that has led to the increased reliance on molecular diagnostics, they could not be effectively diagnosed even if detected, and would be assumed to be novel pathogens. A similar initiative covering viruses, viroids, and virus-like agents of Vitis has also recently been published (Fuchs et al., 2025).
Each NPPO ultimately decides whether a pest should be regulated or not. Such decisions will also be influenced by the level of risk aversion at a national or regional level. The increase in reports of new viruses impacts upon plant health diagnostics, policies and regulation, but ultimately on the trade of goods. To make well informed, risk-based decisions, information on biological relevance, epidemiological and contextual information is required. This knowledge gap expands as more new virus findings arise but the limited resources to acquire and publish the relevant information cannot match the pace of discovery (Hou et al., 2020). Therefore, there is a requirement to investigate alternative methods to provide risk assessment data.
Recommendation 4: Reduce the biosecurity risks through inclusive and equal access virus research and technology
With the rapid adoption of HTS technologies in many countries over the past decade, it has become apparent that the inequalities which exist between communities is affecting the global knowledge of risk from emerging and newly described plant pathogens. Some communities have greater engagement with, or educational and financial access to, research and technology than others. Here, we argue for purposeful and targeted actions to achieve equality of inclusion and access both locally and globally. These equalities will create increased harmonisation between community members, researchers, risk analysts, biosecurity authorities, and policy makers at both national and international levels. Such balanced access and connectedness of people and systems will enable a much needed step change to reduce biosecurity incursions of phytopathogenic viruses. In the short term an increased knowledge and resource capability may result in an increase in first reports and virus discovery. However, in the medium to long term this will result in safer movement of plant products and stable market access of plant commodities.
While even the best researched biosecurity systems are not “fail safe”, fewer resources for research inevitably lead to poorer outcomes. Community-, jurisdiction-, country- or regional-wide equalities around research and technology are derived from multiple drivers, including historic, political, financial or geographic, which result in the current range efficacies of border biosecurity worldwide. While ‘One Health’ (WHO, 2024) recognizes that the health of life on earth, including humans, animals and plants, is closely linked and interdependent and ‘One Biosecurity’ (Hulme, 2020) recognises the need for an interdisciplinary approach to biosecurity policy and research across human, animal, plant, and environmental health, neither sufficiently addresses the need for inclusive and equal access to research and technology. One Health and One Biosecurity work well when they engage across the spectrum of financial, educational and access well-being. As we work towards a zero-hunger world and aim for better lives for all, it is essential that plant health continues to play its vital role in the nexus between human, animal, plant and environmental health.
The historic and extant inequities between communities, regions and/or countries of engagement and access to virus research capability are major contributors to biosecurity risks. The CARE principles were developed by the Global Indigenous Data Alliance to support Collective benefit; Authority to control; Responsibility; and Ethics (Carroll et al., 2020). The differences in support for research capability result in extreme disparities of plant virome description and biological understanding that result in heightened biosecurity risk, especially when coupled with trade (Figure 1 and 2). All communities would benefit from increased inclusivity and access to virus research and technology. Here we examine two potential changes that may provide benefit to all involved; levelling the playing field through purposeful complementary teams that bridge access to research and use as a guidance the CARE principles to bridge the technology gaps.
Levelling the playing field through purposeful complementary teams that cross the critical divides of research and technology access
Inequalities worldwide are measured by a range of approaches and each measure reveals different equality gaps in involvement and access to research and technology. This invariably highlights distinct actions that can be taken to bridge the identified access gaps. Developed and developing countries have been termed Global North and Global South based on socio-economics and politics (Odeh, 2010). This splits the world into two with the ‘North’ comprising rich (high gross domestic product, GDP) developed and mostly temperate countries, and the ‘South’ comprising poorer and typically former or currently colonised countries nearer the tropics. Higher national wealth, e.g. GDP per capita, can fund more research as seen in the Global North by comparison to the Global South. Research communities require greater focus on connection and collaboration across the national wealth spectrum to redress this imbalance.
In the search of solutions to facilitate connection and cross-collaboration between the Global North and South, and to improve coordination at local and global levels, a conceptual model of a global surveillance system for crop diseases has been proposed (Carvajal-Yepes et al., 2019). This system recognizes the importance of diagnostic networks in strengthening the capacity of NPPOs that have limited access to reagents, facilities, or trained staff, helping to reduce biosecurity risks through inclusive and equitable access to research and technology. Efforts to identify specific regional knowledge gaps on diagnostics and surveillance have been undertaken by CGIAR in the Global South, aiming to plan context-based capacity strengthening and provide technical expertise to support national institutions’ surveillance efforts (Carvajal-Yepes et al., 2022). The project CONNECTED examples the purposeful and successful establishment of a multidisciplinary research community formed across equality gaps, in this case around the topic of African vector-borne plant viruses (Ockendon-Powell et al., 2024). Establishing networks to build and enhance understanding and capacity in HTS is a crucial first step in accomplishing this recommendation (4), especially since NPPOs have shown interest in implementing this approach for routine diagnostics. Inclusion of NPPOs, diagnosticians and researchers from both the Global North and Global South ensures equal access to research and technology and addresses a wide range of issues such as biodiversity of crop and their co-evolved viruses as well as the impact of plant viruses on indigenous plants due to spill over (from exported plants to newly encountered plants) or conversely on crop plants due to spillback (i.e. “new” viruses from indigenous plants to imported crop plants.
Use the CARE principles as a guide
The systems in place in the Global North either share data on public databases (e.g. National Center for Biotechnology Information, NCBI), or patent, or protect intellectual property associated with nucleic acid sequence datasets. While undertaking HTS to identify viruses, virologists (or other researchers) often generate sequence data of the viruses as well as their host. In accordance with CARE principles, some guidelines for researchers to consider have been compiled to assist a strategic shift towards Indigenous rights being recognised and empowered within plant pathology, internationally. These guidelines address: how to plan a project or collect endemic/native samples; who are the people necessary to consult, include and compensate on a project; how to incorporate and elevate Indigenous science and knowledge; and what methods to use to disseminate your research” (Ehau-Taumaunu et al., 2024). The principles of CARE apply to Indigenous communities, as exampled with an endemic virus (Rabbidge et al., 2021), and to all communities as exampled by the CONNECTED project (Ockendon-Powell et al., 2024).
Together, we can reduce biosecurity risks from phytopathogenic viruses through projects that develop and deliver respectful and mutually beneficial collaboration. Additionally, these projects may represent how a cultural perspective of a particular group of Indigenous peoples can complement the interpretation of research. Practising the CARE principles strengthens thoughts and actions that create the TOGETHER that is required for biosecurity. These principles complement the level playing field so that all people, societies and the biodiversity, knowledge and data are equally acknowledged and respected.
Inclusion of Global South researchers in conferences and workshops facilitates the formation of collegial networks which can initiate inter-regional projects, share facilities, techniques, and data, and potentially improve the health and prosperity of communities. The ultimate goal is to establish a geographically balanced baseline of information representing plant/virome origins and diversity. Coupled with biological knowledge, such a global plant virome would underpin optimal biosecurity decisions about risks and management options.
Conclusion
Over the last decade, it has become apparent that the development of HTS as one of the frontline biosecurity tools requires researchers and plant protection agencies to go beyond the development of techniques and approaches to begin to address more fundamental questions regarding the scope and purpose of plant health diagnostics. However, there is a rapidly developing gap in access to technology between the better-resourced regions and under-resourced ones, particularly in sub-Saharan Africa, South and Central America, and South and East Asia. We note several exceptions to this observation in the Global South, where several “comparatively” well-resourced facilities established in national and regional programs, and international organizations such as the CGIAR laboratories were established with special project funding. Another challenge in the Global South is access to reagents for HTS due to a lack of distributors, prohibitive costs (offered at full price, not at discounted rates provided to universities in Europe and the USA by the manufacturers), and high import duty (on par with commercial imports) in many countries, all of these factors further reduce the affordability of using the technology even in well-resourced laboratories. This imbalance in technology access and funding creates an “uneven playing field” which will ultimately restrict the use of the technology where it can be of greatest benefit. This situation presents a challenge for plant health, with some countries now having over a decade of familiarity with HTS diagnostics, bioinformatic and analytical pipelines, etc., while others still aspire to implement them. This discrepancy in experience could impact trade through erroneous regulation of recently discovered viruses, which may be common and widespread. Frameworks developed to support regulatory decision-making (Massart et al., 2017, Fontdevila Pareta et al., 2023) should be assessed for their applicability in resource-poor environments.
Plant health regulation is based on species lists which in turn are based on literature reviews of biological characterisation data. This approach lacks the flexibility to handle the increasing number of new detections, increasing the gap between science and risk-based policies. On such regulatory lists are an increasing number of organisms, such as many viruses belonging to the genus Begomovirus (> 400 species), that have been assigned a blanket quarantine status in the EU, the United Kingdom and Switzerland (EPPO/OEPP, 2024). A large proportion of these species have been given this status without specific knowledge of their geographic distribution, host range, or the potential risks posed to the EU region. The effect of this approach, in combination with the use of new technologies in diagnostics such as HTS is two-fold: there is an increasing number of regulated virus discoveries that may require action while there is also an increase in novel viruses with often limited knowledge necessary for well-informed risks assessments (Fox et al., 2019, Silvestre et al., 2020, NVWA, 2022). In a general sense, it is therefore desirable to not only rely on the ‘detection of a sequence’ when assigning a regulatory status, but also to include the relevant contextual information such as biological and geographical properties of the organism, in order to support a risk assessment and deploy scarce resources more effectively (Fontdevila Pareta et al., 2023). Ultimately, plant health policies will be impacted by the use of HTS and while this represents a key challenge, it is also an opportunity for a future state of knowledge-driven plant health protection.
In regulatory settings, to ensure maximum confidence of the diagnostic analyses for virus presence, at least two independent methods are recommended, preferably utilising different biological principles i.e. if initial detection is by a nucleic acid based method (PCR/sequencing) then a confirmatory test should ideally use a different diagnostic target such as serology or bioassay (EPPO/OEPP, 2018a). When diagnostic tests lack specificity or sensitivity, additional tests or additional evidence are often required. However, with many HTS tests, complete structurally and functionally annotated virus genome sequences can be obtained. Therefore, HTS may, in some instances, be used for simultaneous detection and identification, without the need to resort to a secondary method for confirmation. Recently plant health laboratories are describing validated HTS tests performed under ISO 17025 accreditation for virus screening and/ or identification (EPPO/OEPP, 2022). However, in critical cases (e.g. where plant health action or trade restrictions may result from the diagnosis, or on finding a novel virus) a secondary confirmation, utilising an alternate biological principal, may still be desirable. Largely, the outcomes of the diagnostic HTS pipeline are heavily reliant on the diagnostic expertise of both bioinformatics specialists and virologists who interpret the contextual information. Hypotheses regarding putative causal agent(s) should be provided within the context of the plant sample. Therefore, information on the sample’s origin, processing (e.g. washed/unwashed), host, symptomatic status, and other relevant details becomes crucial.
Understanding the (relative) importance of putative findings is crucial in facilitating plant health risk assessment and in prioritising plant health actions, including delivering information supporting delisting or listing as a quarantine pest. To gain these insights, sharing complementary observations among laboratories and countries is necessary and has been demonstrated to efficiently provide context to novel findings. Besides closing the knowledge gap, this will also help in unnecessary duplication of research efforts that strains budgetary resources.
Data sharing has helped to determine the distribution and host range of plant viruses and in at least one case has prevented unnecessary regulatory listing. of Actinidia virus X, was being considered for regulatory listing within the EU, pre-publication data sharing indicated that a historically described species, plantain virus X, was synonymous with this virus, and was already widespread in the EU (Hammond et al., 2021). For the emerging virus Physostegia chlorotic mottle virus, similar data sharing helped to reveal the range of hosts, and its geographic distribution (Temple et al., 2022, Temple et al., 2024). These promising examples originated through organic discussions between research teams, and attempts have been made to take a more formalised approach (Euphresco, 2020). This approach has the objective of sharing high level meta-data of unpublished sequences with partner labs (not the genomic data itself). This creates an atmosphere of trust, avoids issues of confidentiality and access and allows direct partner-to-partner data sharing to be investigated in an efficient way.
There are however various constraints in exploiting data sharing including time availability and prioritisation, uncertainty regarding ownership and sensitivity of data, and concerns about unauthorised exploitation (see CARE principles). Another constraint is with whom the data is shared. The willingness to share data partly depends on the provider’s trust that it will not be misused or disclosed without authorization. Frequently, such data-sharing is often through direct partner-partner sharing that is initiated through ‘chance encounters’ as described above. The constraints of data sharing, combined with limitations of these partner-to-partner working, results in the high amounts of data generated not being (fully) exploited.
Whilst the current use of NCBI as a public repository for sequence data allows interrogation of what sequence has been reported, this outlet lacks key contextual data such as the rationale for the initial detection, and without this context the significance of such findings cannot be easily verified. Challenges to these more formal approaches have included time to collate and manage the sharing of data, and finding the right level of data to share to protect both intellectual property and prevent unnecessary trade regulation. To try and further enhance this type of collaboration we recommend that national, regional, and international initiatives include an open approach to plant health sequence data sharing within future projects. Other next steps which should be explored to safeguard privacy include routes for sharing decoupled sequence data or metadata for initial peer-to-peer collaborative sharing, as well as evaluating more formalised approaches for sharing of data between plant health research groups.
Within some regional plant protection organisations, such as EPPO (European and Mediterranean Plant Protection Organisation), guidelines and standards have been published to help harmonise the implementation of HTS across the region (Lebas et al., 2022, EPPO/OEPP, 2022, Massart et al., 2022). However, even within this region there is uneven access to research and diagnostic funding, and regional coordination of plant health research including sharing of knowledge and experience has been highlighted as a key priority (Giovani et al., 2022). Expanding such a research coordination approach onto a global footing would help to address some of these issues, and is an area where the IPPC and Euphresco are already working to expand the work beyond the current Euphresco member countries (IPPC, 2023, MAFF, 2020). The potential for HTS to be used as part of a global plant health surveillance system, giving early warning of future food security threats, should not be ignored. Whilst the issues discussed here are primarily focused on viruses, this is a consequence of these challenges and the new detection approaches being most acutely felt in plant virology. However, it is anticipated that, in time, similar challenges are likely to also affect other disciplines, and especially those concerned with obligate phytopathogens such as phytoplasmas and liberibacters.
The insights laid down by MacDiarmid et al. (2013) still resonate meaningfully a decade later despite significant technological advances. While some details or emphases may have shifted slightly, the need remains for (1) countries to increase baseline surveillance, (2) standardized nomenclature for sequence data of putative novel viruses to differentiate from characterised viruses, and (3) a greater focus on fundamental biological research to support risk assessments in light of the deluge of new discoveries from HTS methods. Additionally, the disparities observed in this new decade highlight the new recommendation to (4) work towards a global level playing field with equal inclusion and access to research and technology. This equality will create increased cooperation among community members, researchers, risk analysts, biosecurity authorities, and policy makers at national and international levels, leading to a significant reduction in biosecurity incursions of phytopathogenic viruses.
Acknowledgements
The authors would like to thank Darren Snaith for his artwork (Figure 1), as well as Rebecca Gough and Nick Waipara for their review prior to submission. Preprint version 3 of this article has been peer-reviewed and recommended by Peer Community In Infections (https://doi.org/10.24072/pci.infections.100239; (Schumpp, 2025)).
Funding
The authors declare that they have received no specific funding for this publication and the work was prepared during their work for their respective institutes.
Author Contributions
Initial conceptualisation was from Adrian Fox, Marleen Botermans, Robin M. MacDiarmid, and Brendan Rodoni. All authors contributed equally to further development of the conceptualisation, writing and reviewing the original draft.
Conflict of interest disclosure
The authors declare that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article. The authors declare the following non-financial conflict of interest: one author is a recommender for PCI Infections.
Data, scripts, code, and supplementary information availability
Data supporting Figure 2 are available online: https://doi.org/10.5281/zenodo.14013362 (Fox et al., 2024).


 CC-BY 4.0
CC-BY 4.0