Introduction
There is growing demand for biodiversity indicators from international unions, national governments, local management bodies, and corporate and industry actors. Indicators should ideally represent a wide range of biodiversity’s states and functions (e.g. Essential Biodiversity Variables, Pereira et al., 2013; Jetz et al., 2019), yet the development of suitable indicators for certain attributes, such as species abundance and demography, has been more difficult than for others (Schmeller et al., 2018; Waldock et al., 2022). This is at least partially due to challenging requirements regarding spatial scales of useful biodiversity indicators. On one hand, indicators need to be representative at large geographic scales, for example, in the context of countries’ reporting towards biodiversity targets (e.g. Feld et al., 2009). On the other hand, indicators also ideally have good spatial resolution, as the scales relevant for local-level management and planning are often much smaller (Stevenson et al., 2021). This latter requirement is particularly crucial for infrastructure development strategies and for species management and conservation, both of which tend to require knowledge on species abundance and population dynamics (i.e. demographic rates) that is relevant for county- or municipality-level decision making (Christie et al., 2020). Another reason why abundance and population indicators ideally come with good spatial resolution is that there can be substantial amounts of variation in population dynamics and life history of species across space (e.g. Robinson et al., 2014; Horswill et al., 2019). Such variation can arise from differences in ecological contexts (including local habitat and weather conditions, hunting pressures, and interspecific interactions, e.g. Nilsen et al. (2009); Bond et al. (2021)) and needs to be accounted for when developing sustainable strategies for area use, harvest management, and species and biodiversity conservation (Williams et al., 2002).
While large-scale, spatially-explicit indicators for species abundance and populations are clearly needed, development and practical implementation are greatly limited due to the reliance of such indicators on the availability of data from large-scale, long-term monitoring programmes (Proença et al., 2017). Consequently, many countries have been working on setting up, maintaining, and improving such monitoring programmes over the last few decades. Many now well-established programmes focus on breeding birds and butterflies, and examples include the North American Breeding Bird Survey (https://www.pwrc.usgs.gov/bbs/), the PanEuropean Common Bird Monitoring Scheme (https://pecbms.info/), the UK Butterfly Monitoring Scheme (https://ukbms.org/), the Game and Wildlife Conservation Trust Partridge Count Scheme (Aebischer & Ewald, 2010), and the Swiss Biodiversity Monitoring program (https://www.biodiversitymonitoring.ch/).
There is a natural trade-off between quality and quantity of data that can be collected in any monitoring programme: collecting high quality data in a structured manner is costly, requires trained specialists, and hinges on a sufficient degree of top-down control of the programme. This often limits the amount of data that can be collected, while participatory monitoring, i.e. the collection of ecological data by members of the public (also called citizen or community science, Fraisl et al., 2022), allows to greatly reduce costs and extend spatial and taxonomic scales of monitoring at the expense of data quality and risk of bias (Johnston et al., 2023). Consequently, many large-scale monitoring programmes are often limited to presence(-absence) or very simple count observations, making them suitable for the development of indicators of species distributions and perhaps population trends, but usually not of abundance, population dynamics, and demographic rates (Dickinson et al., 2010; Johnston et al., 2023). The exception here are monitoring programmes that succeed in making use of a large number of volunteers that have been trained to collect data and record metadata in a structured manner and according to a carefully designed protocol. For example, in the United States hunters participate in the collection of bands and wings from harvested American Woodcock (Scopolax minor) to estimate survival and age ratios (Zimmerman et al., 2010). At the European level, the recently established initiative “European Observatory of Wildlife” is offering common field- and analyses protocols and aims to establish a network of “observation points” for monitoring wildlife populations at the European level (https://wildlifeobservatory.org/). In Norway there is a monitoring programme for terrestrial game bird species called “Hønsefuglportalen” (https://honsefugl.nina.no/Innsyn/en, = “game bird portal”). It is a line transect survey programme carried out annually in >120 localities across the country (>2000 transects) by trained volunteers using pointing dogs. The programme has a well developed protocol for recording bird observations, auxiliary data, and relevant metadata and established routines for quality control and annual releases of publicly available data via the Global Biodiversity Information Facility (GBIF). As such, it is particularly well suited to become part of a workflow for producing and updating abundance and population indicators on an annual basis.
The line transect data from “Hønsefuglportalen” has been used previously for estimating abundance trends of willow ptarmigan (Lagopus lagopus) across Norway (e.g. Bowler et al., 2020; Nilsen & Rød-Eriksen, 2020), and to test a range of relevant ecological hypotheses (Breisjøberget et al., 2018b; Bowler et al., 2020). However, large-scale estimation of demographic rates underlying abundance trends has thus far remained an untapped potential of the dataset. Nilsen & Nater (2024) recently developed a novel integrated distance sampling model (IDSM) which successfully uses the age of individuals detected along line transects data coupled with radio-telemetry data to jointly estimate abundance, survival, and recruitment across years. In this study, we adapt and extend the model of Nilsen & Nater (2024) to run not just on a single site but on all areas with publicly available line transect data from “Hønsefuglportalen” simultaneously. Unlike several previous studies applying integrated models for population dynamics to multiple (sub-) populations separately and comparing results (e.g. Robinson et al., 2014; Nater et al., 2023), we opt for an approach explicitly integrating across space, thus allowing for sharing of information across locations and – in effect – space-for-time substitution (e.g. Horswill et al., 2019; Morrison et al., 2022). We then apply the resulting multi-area IDSM to “Hønsefuglportalen” data on willow ptarmigan to estimate population size, age-structure, survival, recruitment, and impacts of small rodent occupancy across 41 reporting districts and 15 years (2007-2021) for this culturally important small-game species. We further embed the modelling workflow in reproducible, semi-automated pipelines that will greatly facilitate the repeated calculation of abundance and population indicators at different spatial scales as new data are added every year.
Methods
Study species
The willow ptarmigan is a tetraonid bird with a circumpolar distribution, mainly inhabiting sub-alpine and arctic ecosystems (see e.g. Fuglei et al., 2020). While the species is currently listed as Least Concern (LC) both in the global and Norwegian Red List of Species, it has undergone rather dramatic declines in abundance in Norway since the turn of the 20th century (Hjeljord & Loe, 2022). The main reasons for the long-term decline in abundance remain unresolved, but the willow ptarmigan is considered a sentinel species that is sensitive to both climate change and land use changes (Storch, 2007; Henden et al., 2017). Moreover, being one of only a handful of bird species that spend the winter in mountain ecosystems in Scandinavia, they are important components of the ecosystem as prey for resident predators, such as the gyrfalcon (Franke et al., 2020). The willow ptarmigan has a relatively fast pace of life (Steen & Erikstad, 1996; Sandercock et al., 2005), and can display substantial spatio-temporal variation in demographic rates (Bowler et al., 2020). Population dynamics are characterized by large inter-annual fluctuations in abundance (Hjeljord & Loe, 2022), and previous research has tied these fluctuations to rodent cycles through shared predators (Hagen, 1952; Bowler et al., 2020). This tight relationship between the breeding success of ground nesting birds and the rodent cycle is known as the Alternative Prey Hypothesis (APH) and has been a central part of research on Fennoscandian grouse population dynamics for many decades (Elton, 1942; Hagen, 1952; Linden, 1988; Steen et al., 1988). In addition, spring weather conditions and phenology is known to have considerable effects on breeding success and recruitment rates (Steen et al., 1988; Eriksen et al., 2023). Across their distributional range, willow ptarmigan are an iconic species with a high cultural value, partly linked to their popularity as game species. The latter means that information about spatio-temporal variation in demographic rates and population dynamics is particularly important in order to design sustainable harvest strategies (Eriksen et al., 2023). In addition, being a sentinel species, the willow ptarmigan is well suited as an indicator species for ecosystem status; in Norway it is included in both the main national biodiversity (Nature Index for Norway, https://www.naturindeks.no/Indicators/lirype; Jakobsson & Pedersen, 2020) and ecosystem condition (Assessment of the Ecological Condition, Framstad et al., 2022) assessments.
Data collection, management, and preparation
Line transect sampling
The line transect survey data were collected through a structured participatory monitoring program called “Hønsefuglportalen” (https://honsefugl.nina.no/Innsyn/en). In the first three weeks of August each year, trained volunteer fieldworkers collect observations of willow ptarmigan and other grouse species (rock ptarmigan Lagopus muta, black grouse Lyrurus tetrix, and capercaillie Tetrao urogallus) along predefined line transects. To increase the detection probability, fieldworkers use pointing dogs to locate the birds. A survey team typically consists of at least two people (one dog handler and one person responsible for following the transect line) and one dog. Often, more than one dog is used for a survey, but only one dog should be used at a time. The transect lines vary in length, but are typically between 1 and 8 km (range: 0.3-16.2 km, median: 3 km). When birds are observed, the exact location of observation is reported, along with the perpendicular distance from the transect line, as well as the age and sex of the birds. An observation typically includes 1 - 12 birds (mean = 5.6), with groups > 1 typically representing one brood (female and or male with young-of-the-year chicks). When the surveys are conducted in August, the chicks of the year are able to fly but can be distinguished from older birds as they are still of smaller body size. Since 2019, most of the data has been collected using a mobile app tailored to the monitoring programme, where the field workers can register and get access to the transect lines allocated to them by the local organizers. Prior to 2019, data were collected on a dedicated fieldwork form, and entered manually in a web portal afterwards. After field data has been registered, it undergoes several steps of quality control carried out by local stakeholders and personnel from the Norwegian Institute for Nature Research (NINA). Surveys are carried out on both public and private land. After an initial embargo period, all data from public land are published and made freely available as a sampling-event data set on GBIF (https://www.gbif.org/sampling-event-data). The published datasets contain both metadata about the transect surveys (survey date, line transect length and location, study area ID, etc.) and bird observation data (species, number of birds of different categories (adult males, adult females, juveniles, and birds of unknown category), perpendicular distance to transect line, exact location, and time of observation). Formally, the data from public land is published as three distinct data sets, one for each of the main public land administrators (Statskog, FeFo and Fjellstyrene, respectively).
Notably, the program is not designed as a centralized national monitoring programme, but rather a collection of local and regional survey programs. All involved survey areas use a common field protocol and data collection model. In addition, the local study designs are reviewed by staff at NINA, and common recommendations for study design are provided. However, because participation by stakeholders is voluntary, the spatial distribution of transects and sampling effort is not homogeneous across space. In general, sampling effort is higher in South-Eastern and Central Norway, intermediate in Northern Norway, and low in Western and Southern Norway.
In this study we used all publicly available data for the period 2007-2021, which included a total of 2225 transects in 41 different reporting districts spanning 9 counties and 50 municipalities. Transects on which no willow ptarmigan were observed during the study period (i.e. species absence likely due to low habitat suitability) were not included. After this initial filtering, a total of 2077 transects were included in the analyses.
Radio-telemetry study in Lierne
The model of Nilsen & Nater (2024) integrated line transect data with radio-telemetry data from an ongoing field study of marked willow ptarmigans in Lierne municipality in Central Norway. The main study area in Lierne is located on public land with harvest management representative for the larger multi-area study presented in this paper. From 2015 to 2019, around 50 birds were captured in winter (late February or early March) each year and fitted with VHF collars. This included males and females, and young-of-the year (8-9 months at capture) and adult (>1 year old) birds. The marked birds were then monitored on a regular basis until they either i) died, ii) their transmitter’s battery stopped working, or iii) we lost contact with the bird for other reasons. For most of the year, the birds were monitored at least once a month by radio triangulation. Most of the fieldwork was conducted from the ground, but to avoid data gaps, the birds were also triangulated from helicopters in May, September, and November. During the breeding and chick-rearing season (May to July) birds were monitored more often, and during December and January we obtained fewer observations due to challenging field work conditions. A proportion of the birds were harvested annually in the regular recreational harvest, and birds that were harvested were reported as shot to the field personnel. In addition, as the collars had mortality switches, we were also able to locate and retrieve a high proportion of birds that died from natural causes, resulting in a known-fate mark-recapture dataset. Several previous studies found no effect of the telemetry devices on ptarmigan survival and further details on the radio-telemetry study can be found in Israelsen et al. (2020) and Arnekleiv et al. (2022).
In this study we used data from years 2015 - 2020, and the total sample size across these years was 139 birds for the Aug-Jan period and 258 birds for the Feb-Jul period. We pooled data for males and females as survival was previously found to be very similar (Israelsen et al., 2020) and did not distinguish age classes for analysis.
Rodent occupancy data
As part of the line transect sampling (see above), observers are also requested to report whether they have seen any small rodents while surveying a transect. For each transect survey, this information is recorded as 1’s (small rodents spotted at least once) and 0’s (no small rodents spotted). We aggregated this data into area- and year-specific rodent occupancy covariates by averaging the 0 and 1 reports for all transect surveys within a given area and year and subsequently z-standardizing values. We note that while we refer to the covariate as “rodent occupancy” throughout the manuscript, it can be interpreted as an index for rodent abundance.
National-scale integrated model
Integrated distance sampling model (IDSM) for willow ptarmigan
Nilsen & Nater (2024) recently developed an integrated distance sampling model (IDSM) which jointly analyses line transect and radio-telemetry data and applied it to willow ptarmigan in the Western part of Lierne municipality in Norway. The model consists of a population model with two age classes (juveniles and adults) and four data likelihoods: 1) likelihood for observation distances from transect lines for estimating detection probability; 2) likelihood for age-specific counts on transect surveys for estimating numbers of juveniles and adults present; 3) likelihood for juvenile to adult ratios observed at the locality level to provide estimate recruitment rate (as juveniles/adult); and 4) likelihood for known-fate telemetry data to estimate seasonal and annual survival. Below, we describe our new extension of this model to include data from several areas as opposed to just one. For more detailed information on the single-site model, including tests of model performance, see Nilsen & Nater (2024).
Multi-area model extension
For applying the ptarmigan IDSM across all 41 reporting districts we included an area index in all model parameters (Figure 1) and enabled sharing of information among areas by explicitly modelling spatial variation alongside shared temporal and residual variation in vital rates and detection parameters.
The spatially-explicit formulation of the two age-class population model can be written as:
-
\(\begin{matrix} D_{juv,x,j,t + 1} & = D_{ad,x,j,t + 1}*R_{x,t + 1} \\ D_{ad,x,j,t + 1} & = S_{x,t}*\left( D_{juv,x,j,t} + D_{ad,x,j,t} \right) \end{matrix}\)
Here, \(D_{juv,j,x,t}\) and \(D_{ad,j,x,t}\) are the densities of juvenile and adult ptarmigan in survey site (= transect) \(j\) of area \(x\) in year \(t\), respectively. Both juveniles and adult survive from year \(t\) to \(t + 1\) with an area- (\(x\)) and year- (\(t\)) specific survival probability \(S_{x,t}\), and survivors produce the next generation of juveniles according to an area- and year-specific recruitment rate (\(R_{x,t}\)).
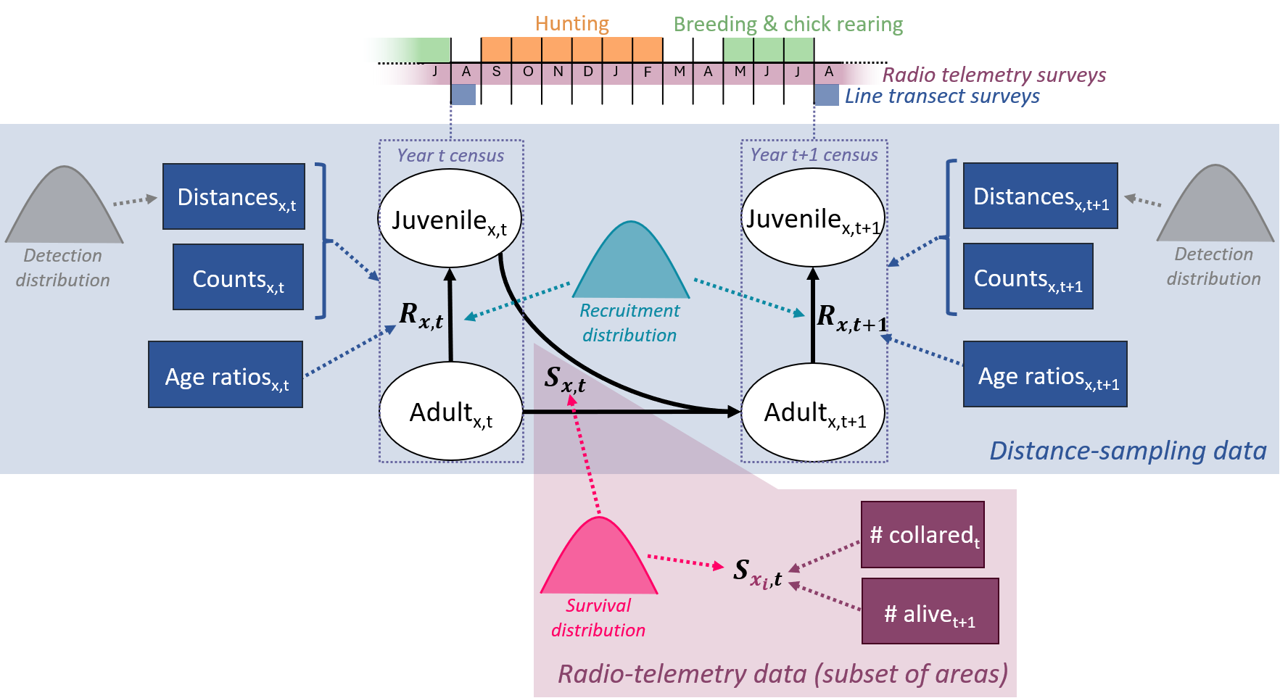
Figure 1 - Graphical representation of the willow ptarmigan life cycle with two age classes and the data sources, as well as joint distributions, included in the integrated distance sampling model. Indices \(t\) denote year while \(x\) represent the added dimension for area (corresponding to 41 reporting districts). Juvenilex,t = juveniles in area \(x\) year \(t\). Adultx,t = adults in area \(x\) in year \(t\). \(R_{x,t}\) = recruitment rate in area \(x\) in year \(t\). \(S_{x,t}\) = survival probability from year \(t\) to \(t + 1\) in area \(x\). Note that the additional site (=transect) dimension, \(j\), is omitted for the sake of illustration.
The initial densities of adults, \(D_{ad,x,j,1}\), are modelled for each site (= transect) as random realizations of log normal distributions with area-specific log means (\(\mu_{x}^{D1}\)) and log standard deviations (\(\sigma_{x}^{D1}\)). Site-specific initial juvenile density, \(D_{juv,x,j,1}\), is calculated from initial adult density as \(D_{ad,x,j,1}*R_{x,1}\). Survival (\(S_{x,t}\)) and recruitment (\(R_{x,t}\)), on the other hand, are assumed to be the same for all sites \(j\) within a given area \(x\) and were modelled as:
-
\(\begin{matrix} logit\left( S_{x,t} \right) & = logit\left( \mu^{S} \right) + \varepsilon_{x}^{X.S} + \varepsilon_{t}^{T.S} + \varepsilon_{x,t}^{R.S} \\ \log\left( R_{x,t} \right) & = \log\left( \mu^{R} \right) + \beta_{x}*rodentOcc_{x,t} + \varepsilon_{x}^{X.R} + \varepsilon_{t}^{T.R} + \varepsilon_{x,t}^{R.R} \end{matrix}\)
The global means,\(\mu\), and the normally distributed spatial random effects, \(\varepsilon^{X}\), represent the equivalent of what is elsewhere referred to as “hyper-parameter distributions” for sharing information on demographic rates across areas (e.g. Horswill et al., 2019, 2021). We also used this same approach for defining the area-specific effects (\(\beta_{x}\)) of local yearly rodent occupancy (\(rodentOcc_{x,t}\)) on recruitment. In addition to spatial variation in survival and recruitment, we also included large-scale temporal variation through random year effects that were shared by across all areas (\(\varepsilon_{t}^{T}\)) and otherwise unaccounted for variation through year- and area-specific residual random effects (\(\varepsilon_{x,t}^{R}\)). Spatial, temporal, and residual random effects were modelled as normally distributed with globally defined (= shared) standard deviations.
The three likelihoods for data resulting from the line transect sampling (observation distances, age-specific counts, and juvenile to adult ratios; see above) were also formulated as spatially explicit, with year- and area-specific distance sampling detection parameters modelled in the same way as survival and recruitment (except the effect of rodent occupancy, Figure 1). For the known-fate telemetry data (and the seasonal decomposition of survival estimated from it), on the other hand, we did not add an additional area dimension as this data was only available for one study area (the Lierne area).
Model implementation
We implemented our multi-area IDSM in a Bayesian framework using NIMBLE version 1.2.0 (de Valpine et al., 2017) in R version 4.4.0 (R Core Team, 2024). For the likelihood for line transect observation distances we used a custom half-normal distribution developed by Michael Scroggie in the “nimbleDistance” package (https://github.com/scrogster/nimbleDistance, see package vignette for specifics). We used non-informative uniform priors for all parameters, but used biologically sensible boundaries where possible. We simulated complete sets of initial values for all model nodes prior to model running and using pre-defined seeds to ensure reproducibility. Using NIMBLE’s standard samplers, we then ran 5 MCMC chains of 200k iterations each. We discarded the first 110k samples of each chain as burn-in, and thinned the remainder by a factor 30, resulting in a final joint posterior containing a total of 5 x 3k = 15k samples (note that high thinning rates were necessary to constrain memory load of the joint posterior, which included 5141 monitored parameters). In addition to the main model run, we implemented a second version of the model that did not use telemetry data to assess the potential impacts of auxiliary data from a single location on parameter estimates.
Follow-up analyses
Post-hoc variance decomposition
Following model fitting, we calculated posterior distributions for the proportions of variance in survival probabilities, recruitment rates, and detection decay explained by 1) spatial variation (\(var_{area}\)), 2) temporal variation (\(var_{year}\)), 3) residual variation (\(var_{residual}\)), and 4) variation in rodent occupancy (\(var_{rodent}\)). To obtain the proportion of variance explained by each of the component, we divided it by the sum of all the components (\(var_{area} + var_{year} + var_{residual} + var_{rodent}\)). The spatial, temporal, and residual variance components were defined as the square of the estimated corresponding random effects standard deviation from the model while \(var_{rodent}\) was calculated as the variance of all area- and year-specific \(\beta_{x}*rodentOcc_{x,t}\) products. This approach for variance decomposition is equivalent to that used by Nater et al. (2018) and inspired by Nakagawa & Schielzeth (2013).
Calculation of additional demographic parameters
For presentation and interpretation of results, we calculated additional key demographic parameters from the model posteriors. First, we calculated area- and year-specific average population densities, \(meanDens_{x,t}\), by averaging \(D_{juv,x,j,t} + D_{ad,x,j,t}\) over all transect lines \(j\) within area \(x\) in year \(t\). We then proceeded to derive area- and year-specific realized population growth rates as \(meanDens_{x,t + 1}/meanDens_{x,t}\). Additionally, we calculated generation time for each area using two different approaches: per-generation population growth rate (Caswell, 2000) and elasticity of asymptotic growth rate with respect to fecundity (Brooks & Lebreton, 2001). For our particular model, the elasticity to fecundity is equivalent to the elasticity of recruitment rate.
Parameter and sampling correlations
We also conducted correlation analyses on the posterior samples to a) check for potential evidence for vital rate trade-offs and/or density dependence and b) assess to what degree the former may be masked by sampling correlation. For a), we calculated Pearson’s correlation coefficients between area-specific time series of estimated vital rate, population density, and population growth rate for each posterior sample. For b), we calculated Pearson’s correlation coefficients between area- and time-dependent survival probabilities and recruitment rates across all posterior samples.
Reproducible workflows with “R targets” and “Nix”
Reproducibility and ease of repeating analyses was a key focus when developing the multi-area IDSM. To that end, we built a function-based workflow (Figure 2) that includes a variety of options for controlling modelling decisions such as the year range of data to consider, the level of spatial aggregation (i.e. reporting district vs. survey locality), whether to model time variation in survival and/or effects of rodent occupancy, whether to run MCMC chains sequentially or in parallel, etc. We then set up three pipelines for executing the workflow that differ in their degree of automation and user interface to meet different needs and resource constraints. The first is a step-by-step R script to be run manually that is suitable for exploration, development, and debugging. The second is a largely automated “R targets” pipeline (Landau, 2021), which allows executions through a single command and maximizes efficiency by keeping track of the “up-to-date” vs. “outdated” status of different steps in the workflow. The third is a pipeline that is run directly from command line, sets up a fully reproducible environment through Nix (Dolstra et al., 2004), and parallelizes the MCMC outside of R using GNU parallel (Tange, 2024). This option avoids a range of issues that can arise with R’s internal parallelization (e.g. processes running even when the parent R session has been restarted, hard to debug, bad error handling, etc.) and is particularly well suited for running on servers and high-performance computing clusters. For more details on pipeline implementations and options, we refer the reader to the vignettes in the GitHub repository: https://github.com/ErlendNilsen/OpenPop_Integrated_DistSamp.
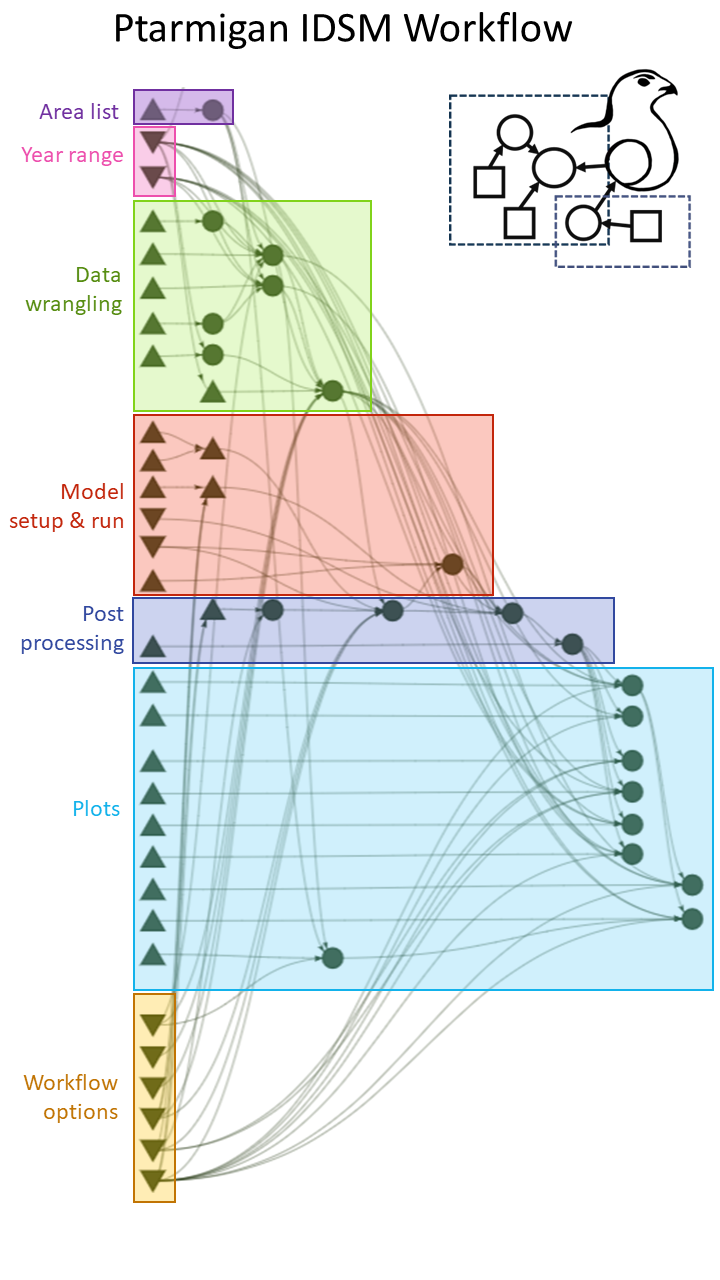
Figure 2 - Simplified graphical representation of the “targets pipeline” for the multi-area modelling setup. Upward facing triangles are functions, downward facing triangles are general options/arguments, circles are objects and outputs (=“targets”) created as part of the workflow. For the full representation, download the repository and call “targets::tar_visnetwork()”. The manual R and Nix/GNU parallel implementations of the workflow have the same structure and built on the same functions and relationships.
Results
MCMC for all central model parameters converged within the given number of iterations, but chain mixing was remained suboptimal even at convergence for some parameters. The inclusion of telemetry data from a single location into the integrated model did not substantially affect parameter estimates beyond the seasonal decomposition of survival in Lierne, where the telemetry data was collected (see online supplementary “Comparison_noTelemetry”). All numerical results in the following are presented as median [95% credible interval] unless otherwise indicated. Posterior summaries (median, 95% credible interval, mean, standard deviation, coefficient of variation) for all main parameters are also provided in the supplementary file “PosteriorSummaries_byAreas.csv”. Supplementary figures (SFs) are provided as .pdf files with captions in “SuppFigures_Captions.txt”; all files are deposited on OSF (https://osf.io/7326r/).

Figure 3 - Median (a) and coefficient of variation (standard deviation / mean) (b) of posterior estimates of average ptarmigan density in the four most recent years (2018-2021) across 41 reporting areas (summarised at the municipality level) in Norway. Darker colors indicate higher median values and higher uncertainty.
Population density
Only during the most recent four years (2018-2021) has data been collected regularly for all reporting areas included in the analyses. During this period, estimated population densities varied between 2.22 [1.56, 3.1] birds/km2 in the area “Statskog og Klinga utm.” close to the coast in central Norway to 55.92 [51.81, 60.03] birds/km2 in “Ålen og Haltdalen Fjellstyre” further south near the Swedish border. In general, recent population density appeared to be lowest in northern Norway and highest in the eastern part of central Norway (Figure 3 (a)). Uncertainty in density estimates was relatively consistent, with a few areas (including the one with the lowest estimated density, “Statskog og Klinga utm.”) sticking out by having substantially less precise estimates (Figure 3 (b)). Populations fluctuated substantially over time in any given area (SF “TimeSeries_popDens1.pdf”) and some years seemed to be indicative of relatively high (e.g. 2011, 2014, 2018) or low (e.g. 2012, 2015) densities across a substantial number of areas.
Population growth rate
Average population growth rates over the last four years (2018-2021) ranged from moderate declines (0.72 [0.64, 0.93] in the “Kongsvoll” area) to > 50% increase (1.55 [1.27, 1.95] in the “Statskog og Klinga utm.” area). In the majority of reporting areas (23 out of 41), populations of willow ptarmigan have been increasing over the period 2018-2021 (Figure 4). Some areas – predominantly in central Norway – also had declining populations, but many of those declines followed upon periods of increase between the start of data collection in 2007 and sometime between 2016 and 2018 (SF “TimeSeries_popDens1.pdf”).
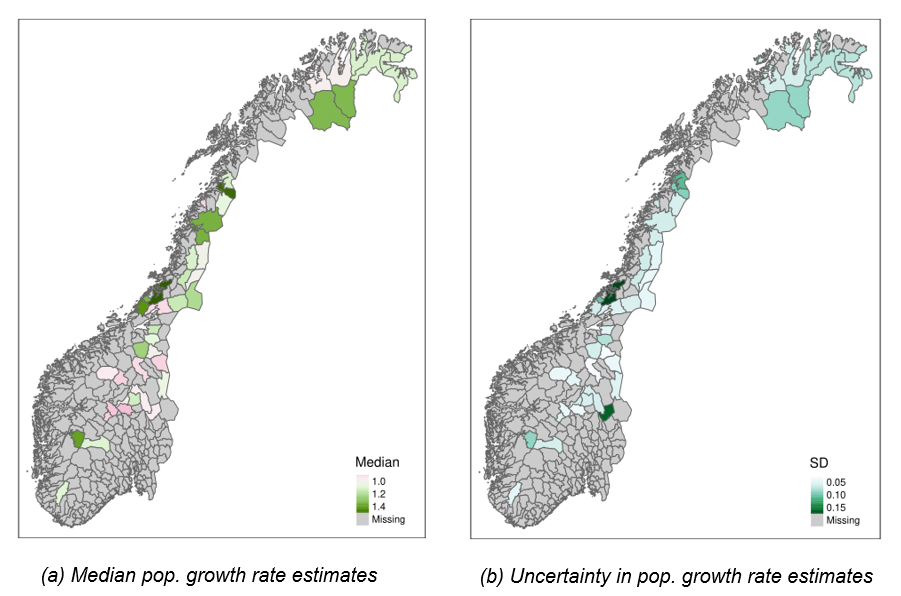
Figure 4 - Median (a) and standard deviation (b) of posterior estimates of average annual population growth rate over the four most recent years (2018-2021) across 41 reporting areas (summarised at the municipality level) in Norway. In a), pinkish colors indicate declining populations while greenish colors indicate growing populations (white = stable populations). In b) darker colors indicate higher uncertainty.
The highest recent population growth rates were estimated for areas with relatively low recent population densities across latitudes but we did not find evidence for a strong association between population growth rates and population densities across areas in general (Figure 5 A). Within areas, however, we found substantial negative relationships between population density and population growth rates, with median correlation coefficients ranging from -0.22 to -0.95 (supplementary file “DD_corrCoef.csv”).

Figure 5 - Posterior summaries (points = posterior medians, lines = 95% credible intervals) of area-specific population growth rate vs. population density over the four most recent years (2018-2021, A), recruitment rate vs. survival probability (B) and rodent effect on recruitment along a longitudinal gradient (C). Color indicates latitude of the midpoint of each area.
Survival probabilities, recruitment rates, and generation times
Annual survival probabilities ranged from 0.33 [0.25, 0.4] (area “Statskog og Klinga utm.”) to 0.43 [0.36, 0.5] (area “Øst Finnmark”) across reporting areas in Norway, with the highest values occurring in the far north and in the mountains in the south (Figure 6 (a)). The global average survival probability across all areas and years (\(\mu^{S}\)) was estimated at 0.38 [0.36, 0.4]. Spatial variation in survival (random effect SD on logit scale = 0.16 [0.06, 0.25]) was relatively low compared to temporal (0.59 [0.4, 0.93]) and residual (0.64 [0.58, 0.71]) variation.
Recruitment rates varied between 1.72 [1.44, 1.98] (area “Gausdal Fjellstyre”) and 3.2 [2.6, 3.95] (area “Statskog og Klinga utm.”) and displayed a spatial pattern opposite to that of annual survival, i.e., lower recruitment rates co-occurring with higher survival rates and vice-versa (Figure 6 (b); Figure 5 B). Across all areas and years, average recruitment rate was 2.4 [2.23, 2.65]. Unlike for survival, the model predicted similar magnitudes of spatial and temporal variation (random effect SDs on log scale of 0.16 [0.12, 0.22] and 0.12 [0.07, 0.19], respectively), and about twice as much residual variation (0.33 [0.31, 0.36]).
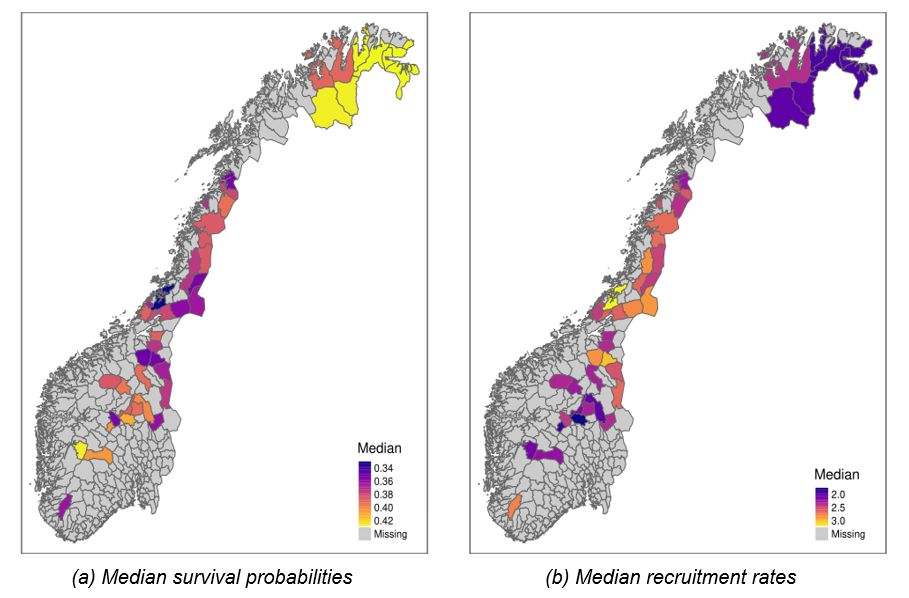
Figure 6 - Posterior medians of average annual survival probabilities (a) and recruitment rates (b) across 41 reporting areas (summarised at the municipality level) in Norway. Darker colors indicate higher median values. Measures for corresponding uncertainty in estimates are visualized in SFs “Avg_pSurv_Map.pdf” and “Avg_rRep_Map.pdf” for survival and recruitment, respectively.
The MCMC chains for many of the area-specific average survival probabilities and recruitment rates, as well as for the global averages for both vital rates, were mixing rather poorly. Despite that, mixing was good and resulting posteriors well defined for the area- and year-specific estimates of survival and recruitment (SF “PostDens_tS_tR.pdf”). There was substantial variation in both vital rates across time (SFs “TimeSeries_pSurv.pdf” and “TimeSeries_rRep.pdf”). In a substantial number of areas, the years 2011, 2014, and 2018 not only supported high population densities (see above) but were also characterized by both high recruitment and low subsequent survival. The overall low density years 2012 and 2015, conversely, often featured lower recruitment and, in some cases, higher survival. Notably, there were also years with very little spatial synchrony, i.e. very different relative yearly survival probabilities and recruitment rates (e.g. 2010 and 2020 for survival and 2013, 2016, and 2017 for recruitment). This same pattern was also reflected in the within-sample correlations between population density in vital rates, which were predominantly positive for recruitment and negative for subsequent survival (supplementary file “DD_corrCoef.csv”). Sampling correlation between annual recruitment rates and survival probabilities was moderate when no time lag was considered (\(R_{t}\) vs. \(S_{t}\), average coefficient = -0.4) and very low when comparing survival to subsequent recruitment (\(R_{t}\) vs. \(S_{t}\), average coefficient = 0.03). Correlation coefficients varied substantially across areas though, featuring both positive and negative values with no clear spatial pattern (SF “SurvRepCorr_Latitude.pdf”, supplementary file “VR_corrCoef.csv”).
Based on estimates of population growth rates and vital rates, we also calculated generation time as both per-generation population growth rate (\(R0\)) and inverse of fecundity elasticity (\(1/elas_{F}\)). The two approaches yielded very similar estimates (median correlation coefficient = 0.95) ranging from 1.31, [1.25, 1.38] to 1.64, [1.56, 1.74] years across areas (SF “GenerationTime_Latitude.pdf”). Spatial patterns in generation time were consistent with those for survival and recruitment, with the highest values occurring in the North and in the mountainous regions in central Norway (SFs “GenerationTime_R0_Map.pdf” and “GenerationTime_elasF_Map.pdf”).
Effects of rodent occupancy
The model predicted a positive global effect of rodent occupancy on recruitment rate (average slope on the log scale = 0.07 [0.01, 0.13]). Nonetheless, spatial variation in the rodent effect was non-negligible (random effect SD = 0.08 [0, 0.15]). This resulted in negative (median) effects in 3 areas, positive (median) effects in 38 areas, and a range of effect sizes from -0.02 [-0.19, 0.1] (area “Selbu Fjellstyre”) to 0.14 [0.02, 0.32] (area “Øst Finnmark”, Figure 5 C, SF “Rep_betaR.R.pdf”). The largest positive rodent effects were estimated for areas in the very North of Norway, as well as in the mountainous regions in the central and southwestern parts of the country (SF “betaR_Map.pdf”). Effects with negative posterior medians were located mostly at intermediate latitudes, but we note that all of these had posterior distributions featuring substantial overlap with 0 (Figure 5 C).
Detection parameters
Detection decay parameters, which determine detection probability in distance sampling surveys, varied across areas from between 68.07 [61.15, 75.78] in “Namskogan Fjellstyre” to 119.97 [108.51, 133.26] in “Engerdal Fjellstyre, resulting in detection probabilities over the transect sites ranging from 0.43 [0.38, 0.47] to 0.75 [0.68, 0.84], respectively (truncation distance = 200 m) . The global average detection decay was 92.19 [86.59, 98.07] (detection probability = 0.58 [0.54, 0.61]), and in general, higher values were more common in the Southern half of the country than the Northern half (SF”Avg_detect_Map.pdf”). Variation in detection over time was modest on average but the degree of temporal changes varied by area, with some areas having nearly constant detection while others showed variation by factors larger than 1.5 (SF “TimeSeries_pDetect.pdf”). The estimated average among-year variation in detection decay (random effect log SD = 0.07 [0.05, 0.12]) was lower than spatial (0.14 [0.11, 0.19]) and residual (0.14 [0.12, 0.15]) variation.
Variance decomposition
The relative importance of different components for explaining parameter variation differed among recruitment rate, survival probability, and detection decay (Figure 7). The largest portion of variation in recruitment was attributed to residual variation (67.5 [55.7, 78] %), followed by spatial (15.7 [9.3, 25.7] %) and temporal (8.4 [3.2, 20.5] %) variation. Rodent occupancy, which contains both a spatial and a temporal dimension, explained 7 [1.4, 13.7] % of the total variation. For survival, there was large uncertainty in the estimated proportions of variance explained by different components. The model predicted similar potential contributions from temporal (44.2 [26, 67.4] %) and residual (52.4 [30.7, 70] %) variation and suggested that spatial variation was only responsible for 3.1 [0.5, 8.2] % of the total variance. The majority of variance in detection decay was attributed evenly to spatial and residual variation at 45.1 [30.8, 60.7] % and 41 [28.4, 54.6] %, respectively. Temporal factors only accounted for 12.5 [5.1, 28.9] %, of detection variation.
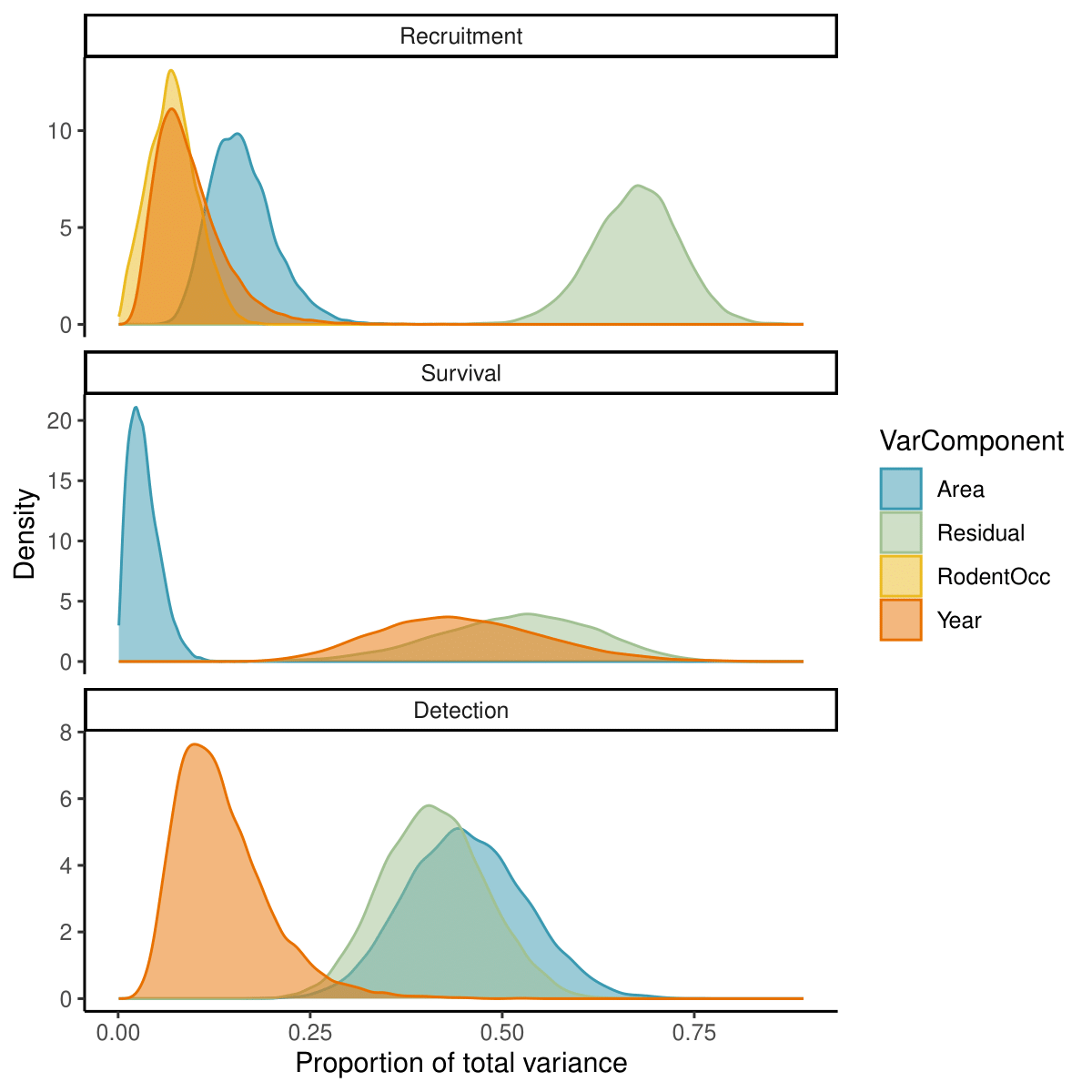
Figure 7 - Posterior distributions for the proportions of parameter variance explained by spatial (blue), temporal (orange), and residual (green) variation, as well as by effects of rodent occupancy (yellow).
Discussion
Building on the work of Nilsen & Nater (2024), we applied a novel integrated population model to data collected through a national-scale participatory monitoring programme to estimate spatial and temporal variation in demography of a culturally important game bird species, the willow ptarmigan. While our study was exploratory in nature, it recovered patterns consistent with ecological and life-history theory including trade-offs between survival and recruitment, and a tendency towards slower life histories at higher latitudes and altitudes. Space-for-time substitution also provided the statistical power necessary for the analysis to provide evidence for the alternative prey hypothesis, i.e. ptarmigans benefiting from high abundance of alternative rodent prey for their predators. Taken together, the results highlight the potential of integrating demographic data across large spatial scales in the contexts of both informing management and creating biodiversity indicators for higher-level reporting.
Abundance and vital rates across space and time
The wide spatial distribution of the line transect monitoring afforded us the opportunity to explore variation in population density and vital rates across a relatively large spatial extent. Ptarmigan densities across the 41 reporting districts included in our analyses varied from around 2 birds/km2 to 55 birds/km2, with the lowest densities occurring far north in the country, as well as on the west coast and in the mountains in central Norway Figure 3 (a). The same spatial pattern was also evident at the level of the demographic rates: consistent with basic life history theory (Stearns, 1992), average recruitment rates were inversely related to average survival probabilities Figure 5, and the slower life histories (higher survival, lower recruitment, and longer generation times) were more common in the northern and mountainous parts of the country. This aligns with previous studies reporting relatively slower bird life histories in alpine / high altitude areas (e.g. Sandercock et al., 2005; Bears et al., 2009; Wilson & Martin, 2011; Alice Boyle et al., 2016). In Norway, the northern and mountainous areas are characterized by more extreme climatic conditions, boasting cold temperatures and short growing seasons. Resulting reduced primary production limits food availability and as ptarmigan are income breeders that use food resources acquired from nesting areas to supply energy and nutrients for egg production and incubation (Sandercock et al., 2005), lower carrying capacity in such areas is to be expected.
We found increasing population trends over recent years in over half of the reporting districts, but population declines were also evident in some areas, particularly in the mountains in central Norway Figure 4 (a). Predominantly increasing population trends are consistent with a recent national-scale analysis by Nilsen & Rød-Eriksen (2020) which found an overall increase in the Norwegian ptarmigan population between 2009 and 2020. While we may speculate that recent population trends could be linked to changes in harvest regulations and/or climatic conditions, considering the whole time-series (2007-2021) illustrated that population densities in all areas were subject to substantial variation across years, featuring periods of stability, increase, and decrease (SF “TimeSeries_PopDens1.pdf”). In most areas, there were also strong year-by-year fluctuations in population density on top of longer-term trends. Some of the resulting “high density years” were highly synchronized across large spatial scales, such as the years 2011, 2013, and 2018. Taking a closer look, we find that these are years that are characterized by high recruitment (SF “TimeSeries_rRep.pdf”), followed by a low survival the year after (SF “TimeSeries_pSurv.pdf”). This often resulted in steep population declines towards the following year. The fact that these same years also match up with observed peaks in rodent abundance in many areas, together with the largely positive effects of rodent occupancy on recruitment estimated by our model (Figure 5 C), provides evidence for the Alternative Prey Hypothesis [APH; Hagen (1952)]. The APH stipulates that high abundance of alternative prey (rodents, in this case) for common predators leads to population growth, and is well-supported throughout the literature for a range of taxa (e.g., Hagen, 1952; Reif et al., 2001; Kjellander & Nordström, 2003), including willow ptarmigan (Bowler et al., 2020). While Nyström et al. (2006) suggested that gyrfalcons, which are specialized ptarmigan predators, do not respond to rodent populations or switch to alternative prey when ptarmigan populations are low, generalist predators, such as red foxes, are likely to shift from preying on ptarmigans to rodents when the latter become abundant (e.g. Breisjøberget et al., 2018b; Bowler et al., 2020). Taking a spatial perspective, the highest latitude and highest altitude areas stood out once more, sporting the strongest effects of rodent occupancy (SF “betaR_Map.pdf”). This could be related to warmer areas generally having larger predator guilds, and consequently more generalists that are able to maintain relatively stable populations irrespective of small rodent abundance (Bowler et al., 2020).
Notably, the conclusive estimation of overall positive effects of rodent occupancy on recruitment in our model was only possible thanks to the integration and sharing of data across multiple areas. When Nilsen & Nater (2024) fit the IDSM to data from only a single area, they were unable to obtain a reliable estimate for the rodent effect due to limited statistical power. Consequently, the space-for-time substitution that comes with extending the model across multiple area allows estimation of covariate effects that otherwise cannot be estimated, and opens up for future possibilities for studying effects of not just rodents, but also other environmental drivers on ptarmigan population dynamics. Doing so may also help with better understanding the mechanisms underlying the large portion of demographic rate variation that could only be attributed to random variation so far. This is the case especially for the relatively large residual variation (Figure 7) but also relevant for constant spatial and shared temporal variation. In previous work based on both marked (Eriksen et al., 2023) and unmarked birds (Novoa et al., 2016; Bowler et al., 2020; Henden et al., 2020), spring conditions have come out as an important predictor of ptarmigan recruitment rates. In general, warmer and earlier springs seem to favour earlier breeding, larger clutch sizes (Eriksen et al., 2023), and resulting higher recruitment rates measured in the late summer and early fall. Bowler et al. (2020) further reported that the strength of this relationship was not consistent in time and space, but was generally stronger in colder areas, similar to what we found for the effect of rodent occupancy here. In practice, measures representing spring conditions, such as the cover of ericaceous shrubs (a proxy for food availability) or spatially-explicit spring green up dates derived from remote-sensing data, thus constitute relevant candidate covariates for future work alongside temperature.
Another important determinant of vital rate variation is density dependence, in particular for exploited species like willow ptarmigan (Andrewartha & Birch, 1954; Willebrand & Hörnell, 2001; Aanes et al., 2002; Sandercock et al., 2011). Negative density dependence has been found in several gallinaceous birds such as northern bobwhites Colinus virginianus (McConnell et al., 2018), Perdix perdix (Bro et al., 2003), and wild turkeys Meleagris gallopavo (McGhee & Berkson, 2007)). For willow ptarmigan, evidence for density-dependent population regulation has been mixed. Myrberget (1988), for example, observed no change in productivity despite a 50% decrease in abundance, while Pedersen et al. (2004) reported strong negative density-dependence over winter and posited that dispersal may be the vital rate that responded to changes in density most strongly. Similarly, Henden et al. (2020) reported negative density dependence when using a Gompertz-model to examine how density and a range of environmental covariates affected willow ptarmigan population dynamics in the northernmost parts of Norway. While we did not explicitly model density dependence in this study, our results can provide some preliminary insights into potential density feedbacks from both a spatial (cross-population) and a temporal (within-population) angle. Comparing average population densities and growth rate across areas did not provide evidence for strong density dependence, but there was a tendency towards the highest population growth rates appearing in areas with relatively low density, and relatively low growth rates in high-density areas Figure 5. When considering density dependence across years within select areas, however, we found that higher density years were associated with higher recruitment the same year, but followed by lower apparent survival probabilities and, consequently, lower population growth rates (as determined by post-hoc Pearson correlation coefficients, supplementary file “DD_corrCoef.csv”). While this seems to support the notion of negative density-dependence, testing for this post-hoc gives results that are confounded with sampling correlation (Freckleton et al., 2006). Our tests showed a moderate degree of sampling correlation between survival and recruitment on average (up to -0.4), but there was substantial variation in the degree of correlation across areas (SF “SurvRepCorr_Latitude.pdf”). Hence, formally modelling density-dependence, possibly using different forms and time-lags, could prove to be a promising extension of our modelling framework in the future.
Implications for management and monitoring
Management decisions made at the resolution of large geopolitical boundaries (e.g., Norway) run a high risk of being inadequate when there is substantial spatial variation in demographic processes and population dynamics, as is the case for willow ptarmigan. In Norway, willow ptarmigan – and small game in general – are managed at the local and/or regional scale, with rather limited national regulation beyond updating the length of the hunting season every fourth year. In effect, management system, regulation type (quota type, season length, number of licences, bag limit etc.), and quota size are governed by the local or regional stakeholders (Eriksen et al., 2018; Breisjøberget et al., 2018a). Thus, while national estimates (abundance and/or temporal trend in abundance) might be important for red listing decisions and for setting the maximum hunting season length, remaining decisions about harvest management are taken locally. The results from our study highlight a large degree of spatio-temporal variation in both ptarmigan densities and demographic rates, suggesting that it is indeed suitable for management decisions to be spatially refined and ideally informed by up-to-date knowledge about recent “local” population processes. Accessible and easily repeatable modelling workflows, such as the one we have developed in this study, can thus become a valuable source of information for local decision-makers.
Our results also provided some insights into the value, and possibly opportunities for improving the monitoring programme. First and foremost, our study demonstrates the tremendous potential within coordinating structured monitoring that employs common sampling protocol, training programmes, and data processing pipelines. These were indeed the prerequisites that allowed us to easily and efficiently integrate data collected across the entire country in a joint analysis, and draw inference on fine-scale spatio-temporal variation in demography and population dynamics at across a large area. While overall less variable across space and time than vital rates, differences in detection probabilities were nonetheless evident (SFs “Avg_detect_Map.pdf” and “TimeSeries_pDetect.pdf”) and may help with mapping out potential for improvement in the monitoring programme. Particularly, we found generally lower detection probabilities in the northern half of Norway. This may be related to habitat features, as the transects in the North might be to a larger extent located in birch forests and rugged terrain, which may hamper detectability. Additionally, the slower life histories in the northern areas are reflected as generally smaller bird clusters as well, and smaller clusters have previously been shown to have a lower detectability than larger ones (e.g. Bowler et al., 2020, see also next section). Our modelling framework can be easily adapted for studying the impact of these and other variables on detectability (see below). Together with ongoing efforts of increasing the number and density of transect lines in Northern Norway, this can contribute to obtaining more precise estimates of both population density and demographic rates, and would strengthen inference particularly in areas with relatively low ptarmigan population densities and less years of data.
Model limitations and outlook
The primary focus of this work was placed on developing an effective pipeline for integrating data and modelling population dynamics across a large number of areas. Consequently, many additional opportunities for improving and refining the modelling framework itself remain. First, the precision and accuracy of model estimates might be increased through better accounting for heterogeneity and potential biases in detection of birds during the line transect surveys. In an earlier study analyzing data from the same monitoring programme, Bowler et al. (2020), found that detection probability was not independent of the size of group birds were part of, resulting in birds in larger groups being more likely to be detected, especially at larger distances. When birds are observed in larger groups, it is also not unlikely that human observers may miscount, i.e. that there is some observation error in the number reported. This could be incorporated by including an additional layer of hierarchy to the observation process (see e.g., Hamilton et al., 2018), and possibly further extended to also account for error in judging the observation distance (e.g., Marques, 2004). Another potential source of bias in our IDSM is related to failure to correctly assign the age class of observed birds. Nilsen & Nater (2024) showed that incorrect age assessment can bias (relative) estimates of survival and recruitment, and while they only found a weak bias in their case study on a single area, the problem may be larger in a multi-area setting that may contain areas with different proportions of misclassified observations. If misclassification happened at random, mixture models could be used to determine the likely age class of individuals to whom no age class was assigned during observations (McCrea et al., 2013). In our case, we might suspect that an observer is more likely to classify an adult bird as juvenile rather than the other way around, and more likely to assign “unknown” age class to juveniles than adults. One reason for this is that observers look for specific signs to classifying a bird as adult (e.g. size, male sound), and might default to juvenile or unknown if the signs are not clearly detected. Future studies should investigate to what degree available information on e.g. group composition could be used for this, and what kind of auxiliary data would need to be collected to reliably model misclassification error.
The second (and perhaps most attractive) aspect of our modelling framework in the context of future work is its spatio-temporal hierarchical structure. While we included spatial, temporal, and residual variation in our framework here, we treated them as independent. Alternatively, spatial (and temporal) correlations among parameters can be modelled explicitly, something that is commonly done e.g. in modern species distribution models (e.g. Pacifici et al., 2017; Guélat & Kéry, 2018). For demographic models, this has rarely been implemented so far, not least due to the fact that few demographic models have sufficient spatial resolution (Schaub & Kéry, 2021). The ptarmigan IDSM presented in this study, however, does have sufficient resolution and our results do indeed support that there is spatial clustering in both overall and time-dependent demographic parameters (e.g. Figure 6, SFs “Avg_pSurv_Map.pdf” & “Avg_rRep_Map.pdf”). Furthermore, we did find that mixing of several of the global and area-specific intercept parameters in the current model was suboptimal, suggesting that there may be much to gain from additional structuring, as well as from development of more efficient MCMC sampling strategies for the resulting extended model. One promising framework for approaching this are conditionally autoregressive models (CARs, Ver Hoef et al., 2018). Such models have been used repeatedly for modelling spatial autocorrelation in species occupancy and demographic rates (e.g. Saracco et al., 2010, 2012; Guélat & Kéry, 2018) and are straightforward to implement using NIMBLE (Lawson, 2020). One possible challenge with using CAR models to explicitly model spatial correlations within our ptarmigan IDSM is that CAR models rely on “neighborhood” relationships between discrete areas and many “neighbors” are missing in our ptarmigan data (e.g. Figure 3). Estimation of latent parameters in missing areas may be possible though (Perry de Valpine, personal communication; Schaub & Kéry (2021) chapter 19), and this may result in a unique opportunity for making predictions of ptarmigan population trends in unmonitored areas, provided that data for a sufficient number and range of areas are available. Here, we may benefit from the fact that the line transect survey data included in this study constitutes just the publicly available part of the data collected through “Hønsefuglportalen” but the programme also includes additional surveys on private land. Extending to data from private land would provide better coverage especially in south-eastern and southern parts of Norway, which includes areas where only very limited amounts of data are collected on public land. Exploring to what degree additional data from “Hønsefuglportalen” could be included in future studies employing an extended IDSM with additional spatial structuring is therefore a worthwhile endeavor.
Finally, including further data beyond the line transect surveys may be relevant in the future, and in particular in the context of informing and improving management of ptarmigan hunting. In the present study, we have used auxiliary radio-telemetry data to supplement information on survival, but since this data was available for only one out of 41 areas, its influence was small. Nonetheless, this illustrates a way for how smaller datasets from single or subsets of areas can be integrated into a large-scale modelling framework. Other relevant data could be included using the same approach, for example data from ongoing nesting success monitoring, data from past studies of marked birds (Sandercock et al., 2011), and data from other monitoring programs for breeding birds based on point counts (see e.g. the Norwegian Breeding Bird Monitoring: https://hekkefuglovervakingen.nina.no/). The most relevant source of data to be included into the IDSM framework in the near future, however, is harvest data. Such data might be available with different spatial and temporal resolutions. First, at the municipality level there are data with national coverage collected annually by Statistics Norway (https://www.ssb.no/). Second, many public land owners have data with much higher temporal (daily) and spatial (harvest area) resolution, including both harvest bags and harvest effort (number of hunters per area per day). As the IDSM framework is, in essence, an IPM, harvest can be modelled through partitioning of survival into cause-specific mortality in the process model and inclusion of relevant harvest data likelihoods (e.g., Gamelon et al., 2021; Nater et al., 2021). While harvest effects on willow ptarmigan have been studied previously, much uncertainty remains (Willebrand & Hörnell, 2001; Aanes et al., 2002; Pedersen et al., 2004; Sandercock et al., 2011). For example, little is known about how harvest pressure and density feedbacks interact on different temporal and spatial scales (Kvasnes et al., 2015), despite this knowledge being crucial for preventing over-exploitation and ensuring sustainable harvest (Williams et al., 2002; Breisjøberget et al., 2018a). Additionally, harvest effects often interact with other (emergent) factors such as climate change and habitat degradation, making predictive models that account for harvest alongside other mechanisms invaluable for informing policy changes (Gamelon et al., 2019).
Reproducible workflows for a sustainable future
Producing a transparent and reproducible workflow for the analysis presented here was a central objective in this study. We have done this by setting up a well documented, function-based R workflow that allows (re-)running the complete analysis from downloading the publicly available data to visualizing the results produced by the IDSM (Figure 2) and that can be implemented using pipelines that employ “R targets” (Landau, 2021) and Nix/GNU parallel (Dolstra et al., 2004; Tange, 2024). Modern applied ecology needs research to be published not just as scientific papers, but as reproducible and well documented workflows (Lewis et al., 2018). This is particularly crucial for research that is (to be) closely tied to management and/or used to create biodiversity indicators that are to be reported nationally or internationally, or to be used by industrial partners (Powers & Hampton, 2019). This is both because of the enhanced transparency and credibility provided by openly available reproducible workflows and because of their cost-effectiveness, which allows for more sustainable use of funding in the mid- to long-term. Finally, open and reproducible workflows facilitate collaboration and inclusion of stakeholders in the research process, paving the path for the translational science that is required for society to tackle the the biodiversity crisis (Rubert-Nason et al., 2021). It is our hope that this study can serve as an example of where to start.
Acknowledgements
We thank Mark Hewinson, Todd Arnold, and one anonymous reviewer for constructive comments during the review process. We are grateful to the numerous volunteer observers (and their canine assistants) who collected ptarmigan observations as part of the “Hønsefuglportalen” monitoring effort. Additionally, we would like to thank Bernardo Brandão Niebuhr dos Santos for his help with spatial data visualization. J. A. Martin was supported by a Fulbright Fellowship and the Norwegian Institute for Nature Research, which facilitated this collaboration.
Preprint version 4 of this article has been peer-reviewed and recommended by Peer Community In Ecology (https://doi.org/10.24072/pci.ecology.100676; Hewison, 2024).
Author contributions
Chloé R. Nater: Conceptualization, Methodology, Software, Formal analysis, Writing - Original Draft, Writing - Review and editing, Visualization.
Francesco Frassinelli: Software, Writing - Review and editing.
James A. Martin: Conceptualization, Writing - Original Draft, Writing - review and editing.
Erlend B. Nilsen: Conceptualization, Methodology, Data curation, Writing - Original Draft, Writing - Review and editing, Project administration, Funding acquisition
Ethics Approval
All data on marked birds were collected with highest concern for animal welfare, with authorization from the Norwegian Food Safety Authority (IDs 8477, 15166 and 22919).
Funding
Funding for both data collection and analytic work was provided by the Norwegian Environmental Agency (grant numbers 17010522, 19047014, and 22047004).
Conflict of interest disclosure
The authors declare that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Data, script, code, and supplementary information availability
The raw data from the line transect surveys is deposited on GBIF and can be accessed freely via the Living Norway Data Portal (https://data.livingnorway.no/). The work presented above is based on versions 1.7, 1.8, and 1.12 for the datasets from Fjellstyrene (https://doi.org/10.15468/975ski; Nilsen et al. 2022c), Statskog (https://doi.org/10.15468/q2ehlk; Nilsen et al. 2022a), and FeFo (Nilsen et al., 2022b), respectively.
The auxiliary radio-telemetry data, rodent occupancy data, posterior summaries, and supplementary figures are archived on OSF (https://doi.org/10.17605/OSF.IO/7326R; Nater et al., 2024b).
All code, including the three pipelines, can be found in the project’s repository on GitHub: https://github.com/ErlendNilsen/OpenPop_Integrated_DistSamp. The results presented in this paper were created using version 2.1 of the code (https://doi.org/10.5281/zenodo.13767267; Nater et al., 2024a).


 CC-BY 4.0
CC-BY 4.0